Team:Paris Bettencourt/Delay
From 2012.igem.org
|
Achievements :
|
Contents |
Overview
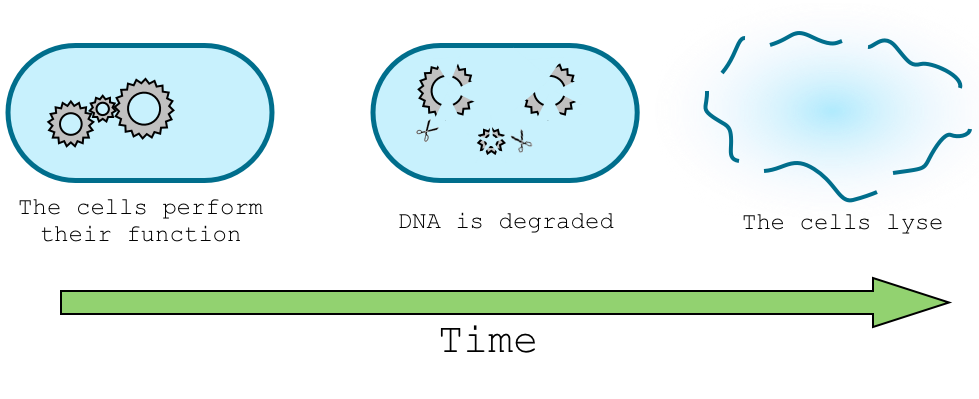
In order to let our genetically modified organism perform its task in the environment, for example sensing nitrate concentration, and in the same time trigger the death of the organism and degradation of its DNA we need to design a delay system that will separate this two task in time. Give a real example of the application.
This part of the design will trigger the DNA degradation mechanism. To do so, we will use two different approaches. The first one, that we called "simple delay system", will be a way to trigger a restriction enzyme, which will cut the plasmid carrying the antitoxin, and thus let the cell sensitive to the toxin produced on the chromosome. In the "sRNA delay system, we use a different strategy. Here, the toxin would be cloned without the Immunity protein, in order to avoid any possible resistance to the toxin due to a non-degraded plasmid. To ensure that the toxin will not be produced while the cells are performing their function, we will use a stationary phase promoter and sRNA repression induced by arabinose.
Simple delay system
Objectives
We wanted to create a simple way to trigger the production of our restriction enzyme, leading to an inevitable death for the cells. The challenges of such a genetic circuit are the following :
- We need to have a long enough delay for the cells to perform their function before activating the "killswitch".
- We need to have a very strong repression (without leakage) of the restriction enzyme while the safety system is not activated.
Design
Characterization
sRNA delay system
Objectives
As an alternative trigger to the «collective suicide» presented in the overview of the project, we tried to build a system in which the toxin would be cloned without the Immunity protein, in order to avoid any possible resistance to the toxin due to a non-degraded plasmid. Such a design comes with several difficulties :
- we need a very strong repression of the expression of the toxin. A leaky expression would harm the cells, and one of the risks is that mutants which do not produce the toxin arise and become selected.
- we need to be able to have a control on the delay, in order for the cells to trigger the safety mechanism only once they have performed their function.
- we need to be sure that the safety system will be triggered at one point. We prioritize the robustness of the delay over its timespan.
Design
We want to use a sRNA as a way to block the expression of the toxin. The concept of our design is described below. Similar systems have been shown to respond in a treshold-linear way [2]
In order to achieve our goal, we started form the construct of Yokobayashi et al. [1] described below.
Even though the cloning is still in process, we hope to build the following construct by the end of the competition.
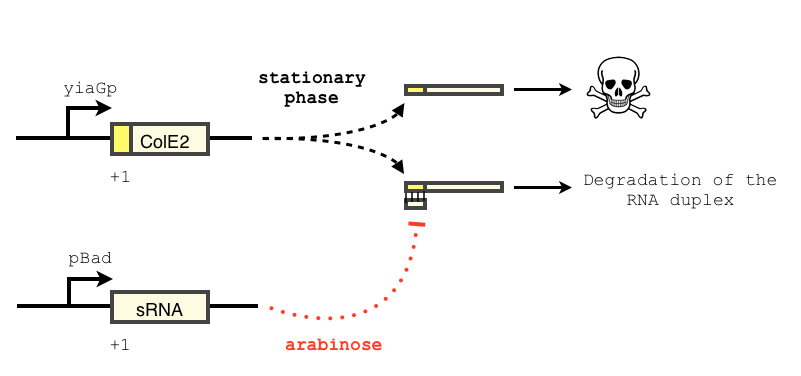
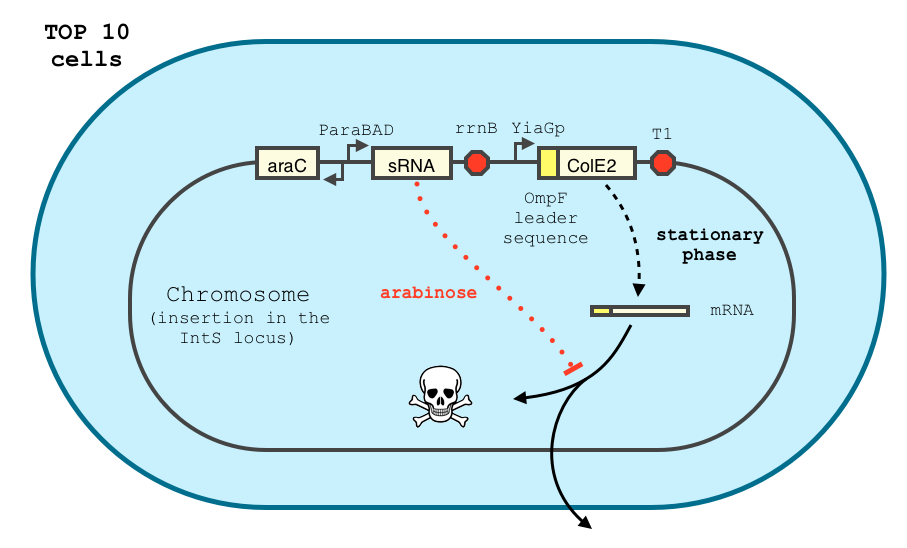
The transcription of the toxin is controlled twice : first, we chose a stationary phase promoter, yiaGp. It is known be recognized by the sigma-S subunit of the RNA polymerase, and not the exponential phase sigma-70 subunit. [3], [4].
We have shown that the yiaGp promoter is indeed activated during the stationary phase in our construct, even though the level of trancription is rather low (see experiments)
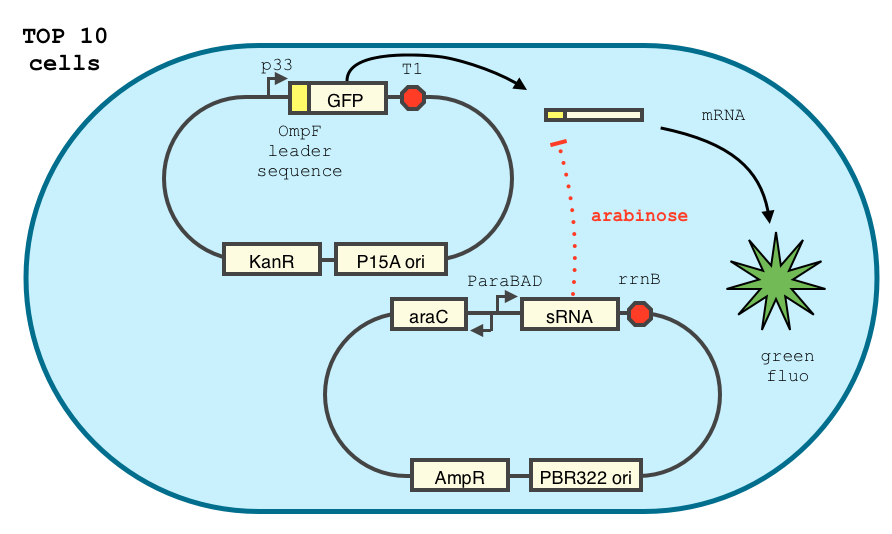
In order to achieve a complete lockdown of the toxin, we added a second repression mechanism, this time at the post transcriptional level. We used and modified the constructs of Yokobayashi et al. to allow a repression of the translation using sRNA [1]. We chose to use the sRNA 7.9, as it has been shown to bind the leader sequence of its target mRNA and not its coding sequence, and to allow a 20 fold repression of the protein’s expression. The transcription of the sRNA is under the control of the pBAD promoter. The coding sequence of colicin E2 is fused to the leader sequence (5’ UTR) of OmpF. The araC protein allows us to use this construct in cells deleted for the arabinose operon (TOP10). We hope that, using this type of cells, the delay will depend on the dilution of the arabinose in the medium as it is not metabolized. We will need to check the effect of extracellular glucose on the level of repression, as the CRP protein is known to repress the pBAD promoter.
Experiments and results
Characterisation of the YiaGp promoter
Using the following construct, we characterized the stationary phase YiaGp promoter in TOP10 cells.
Experimental setup
Results
Present your results
Refined characterization of the Yokobayashi et al. sRNA repression plasmidic device
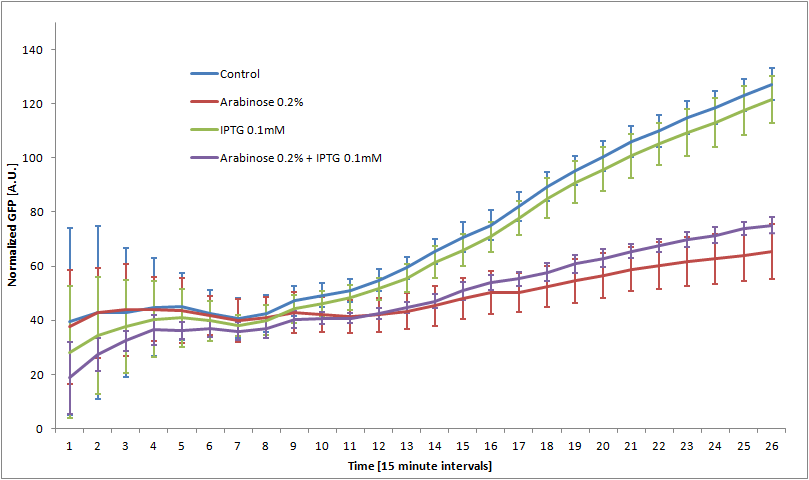
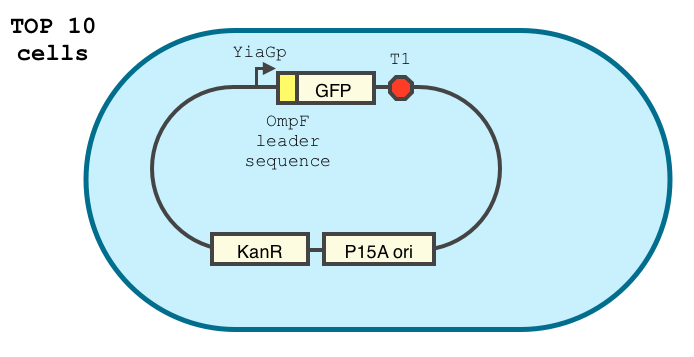
We double transformed TOP10 cells with the pKP33-GFPuv and pBAD-aOmpF-7.9 plasmids (see [1] for details, and click here for a schematic of the genetic system). We chose the 7.9 sRNA because it seemed to show a strong repression of the GFP expression, and bind to the RBS of the aOmpF leader sequence and not to the coding sequence.
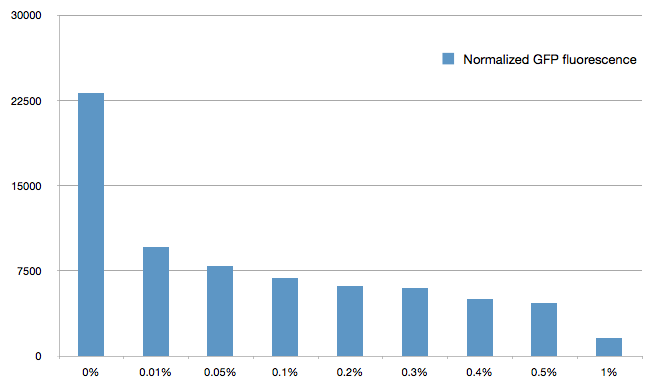
The cells were grown in LB until stationary phase, then diluted 100-fold in LB. After adding various arabinose concentrations to the medium, we monitored the fluorescence of GFP (excitation : 395nm, emission : 509nm) using a TECAN 96-wells plate reader during 10 hours . We had 6 replicates for each concentration. The bar plot below shows the average normalized fluorescence at the final time point for each concentration tested.
Results
Present your results
References
1. Sharma, V., Yamamura, A., & Yokobayashi, Y. (2012). Engineering Artificial Small RNAs for Conditional Gene Silencing in Escherichia coli, 6-13. [http://pubs.acs.org/doi/abs/10.1021/sb200001q| Paper]
2. Levine, E., Zhang, Z., Kuhlman, T., & Hwa, T. (2007). Quantitative characteristics of gene regulation by small RNA. PLoS biology, 5(9), e229. doi:10.1371/journal.pbio.0050229 [http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0050229| Paper]
3. Sharma, U. K., & Chatterji, D. (2010). Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS microbiology reviews, 34(5), 646-57. doi:10.1111/j.1574-6976.2010.00223.x[http://jb.asm.org/content/186/21/7112.long| Paper]
4. Shimada, T., Makinoshima, H., Ogawa, Y., Miki, T., Maeda, M., & Ishihama, A. (2004). Classification and Strength Measurement of Stationary-Phase Promoters by Use of a Newly Developed Promoter Cloning Vector, 186(21), 7112-7122. doi:10.1128/JB.186.21.7112 [http://onlinelibrary.wiley.com/doi/10.1111/j.1574-6976.2010.00223.x/abstract;jsessionid=8C98B1B383242B9DFF45429802F48428.d02t04| Paper]
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment