Team:Paris Bettencourt/MAGE
From 2012.igem.org
|
Aim Removal of four FseI restriction sites from E. coli MG1655 genome. Experimental System Using multiplex automated genome engineering (MAGE) - a technique capable of editing the genome by making small changes in existing genomic sequences. Achievements
|
Contents |
Overview
Multiplex automated genome engineering (MAGE) is a method for large-scale programming and evolution of cells [1]. MAGE is a recently developed technique capable of editing the genome by making small changes in existing genomic sequences. This is accomplished by inserting single-stranded oligos that contain the desired mutations into the cell, and more than one gene can be targeted at a time simply by using multiple oligos. Mediated by λ-Red ssDNA-binding protein β, the oligos are incorporated into the lagging strand of the replication fork during DNA replication, creating a new allele that will spread through the population as the bacteria divide. The efficiency of oligo incorporation depends on several factors, but the frequency of the allele can be increased by performing multiple rounds of MAGE on the same cell culture. Currently, researchers must perform each round by hand, a procedure requiring several hours, but automated processes are being developed to run many rounds of MAGE in succession. Together, these properties make MAGE a highly useful tool for synthetic biology, allowing researchers to easily modify the bacterial genome and generate diversity within a population. We will do it by hand.
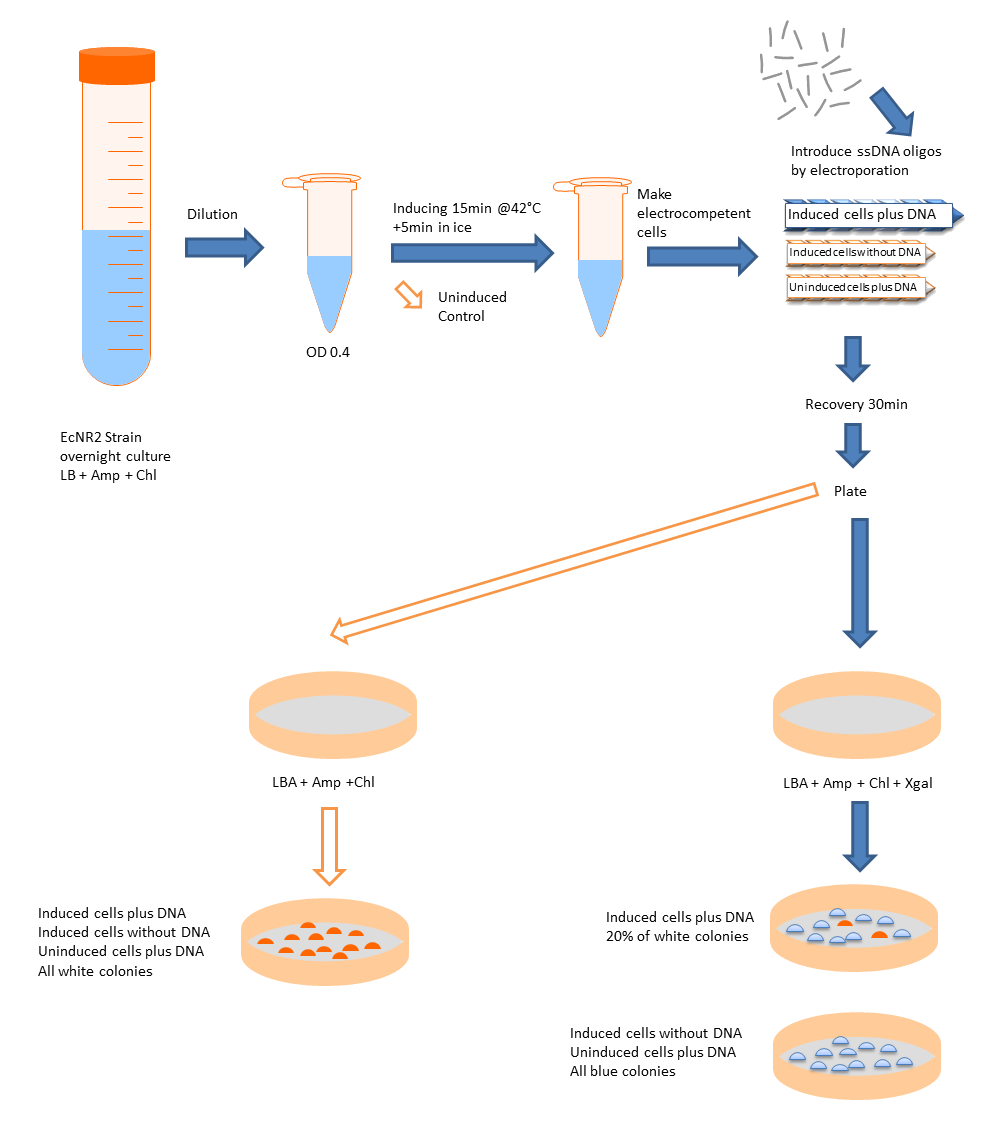
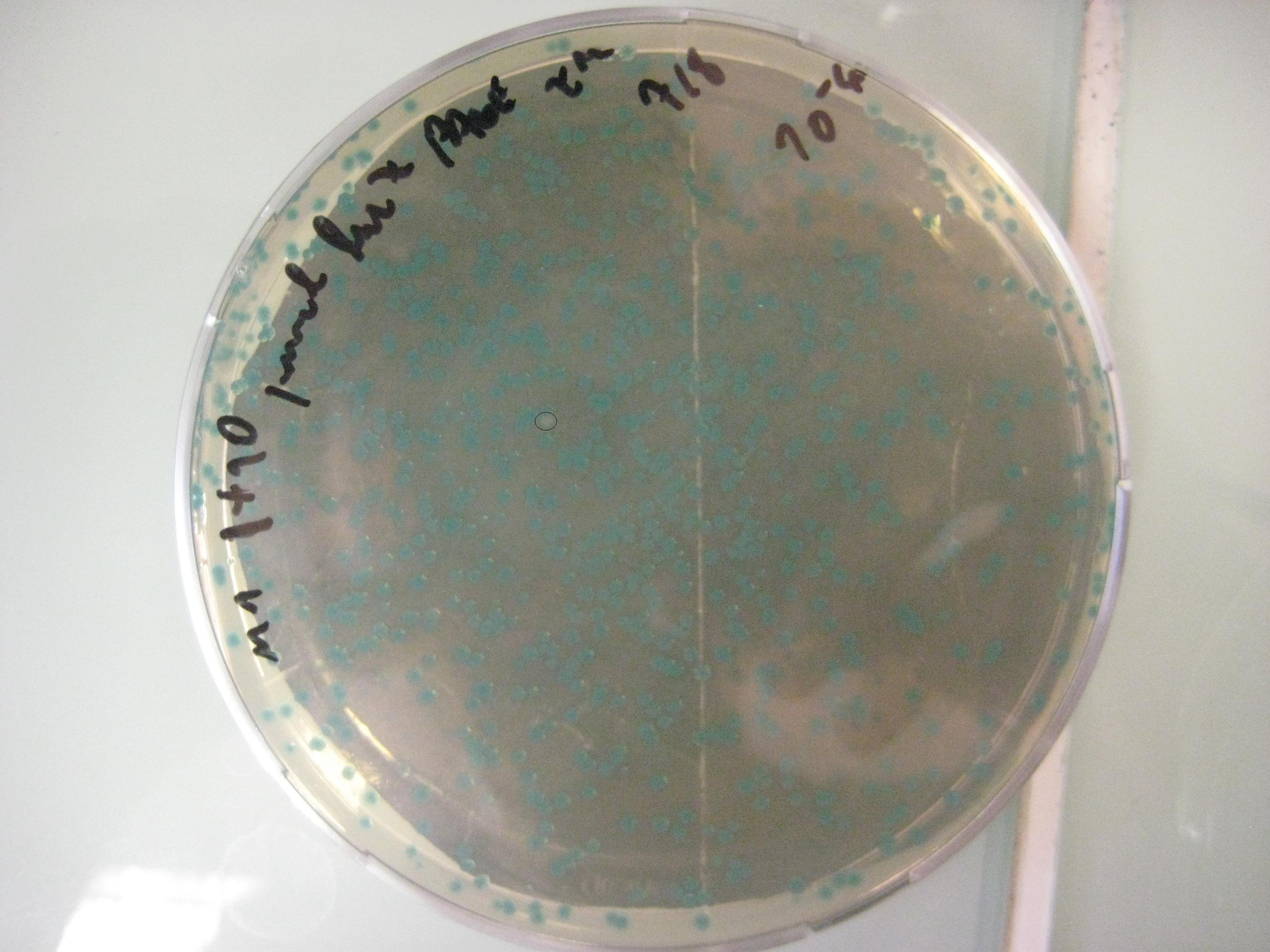
We want to use this method to remove four FseI restriction sites on the E. coli MG1655 genome. By doing this our genome won't be cut by FseI restriction enzyme which we want to express inside of our system. More details can be found in Restriction Enzyme part of the project. To check the efficiency of "MAGE-by-hand" we designed an oligo which introduces a stop codon in the middle of the lacZ sequence. Our mutated strain is easy to screen because the colonies with ineffective lacZ will stay white on an LB plate with X-gal whereas bacteria that were not mutated will have effective lacZ and they will turn blue. Since there is no visual way of screening mutation of FseI sites we needed high efficiency levels. Unfortunately, we never got a satisfactory efficiency level and we abandoned the idea of using this method.
Objectives
- Test engineering of chromosomal DNA using single-stranded oligonucleotides that enter a stop codon in a middle of a lacZ gene
- Delete four FseI restriction sites from E. coli MG1655 genome
Design
Protocol
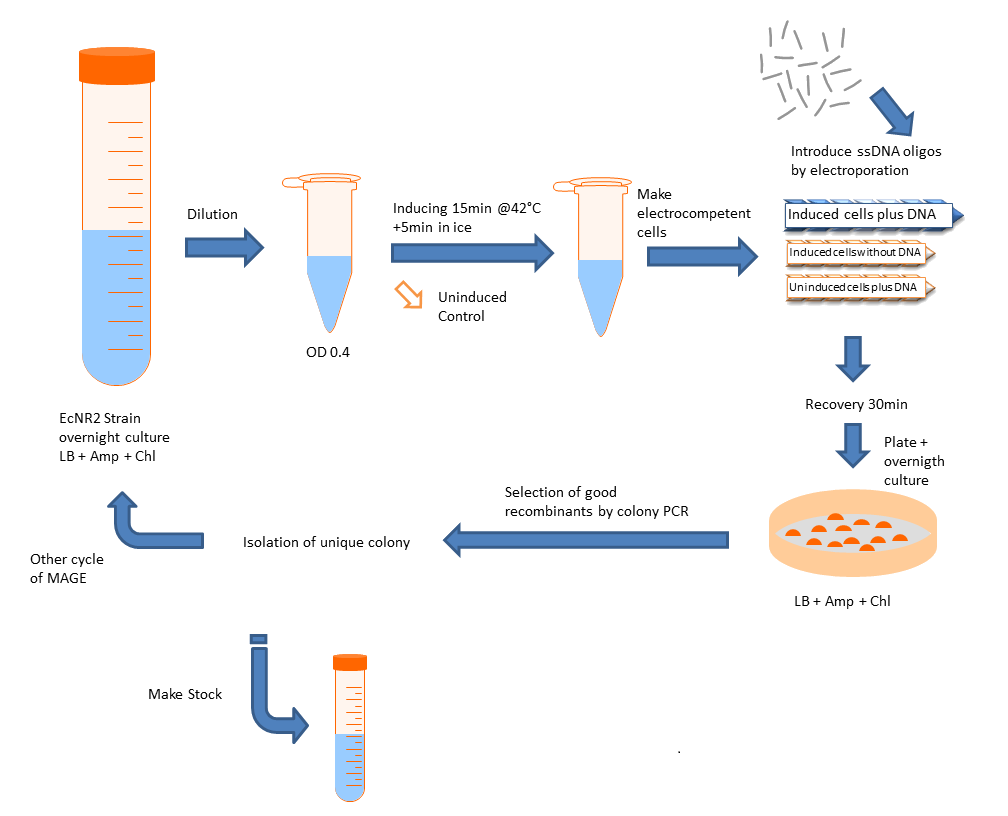
Prepare bacterial cultures
- Inoculate EcNR2 strain from frozen glycerol stock or a single colony into 3 to 5 ml LB medium. Shake at 30° to 32°C overnight.
- Add ~0.5 ml of the overnight culture to 35 ml of LB medium in a 250-ml (baffled) Erlenmeyer flask.
- Place the flask in the 32°C shaking water bath and grow cells at 32°C with shaking for ~2 hr.
- The cells are ready when the A600 is between 0.4 and 0.6. It is important not to over-grow the cells, since stationary phase cells do not express the recombination functions well.
Induce recombination functions
- Transfer half the culture to a 125-ml (baffled) Erlenmeyer flask and place that flask in the 42°C water bath. Shake 15 min at 220 rpm to induce. Leave the remainder of the culture at 32°C; this will be used as the uninduced control that lacks recombination activity. While the cells are inducing, fill an ice bucket with an ice-water slurry.
- Immediately after inducing for 15 min at 42°C, rapidly cool the flask in the icewater slurry with gentle swirling. Leave on ice for ≥5 min. Follow the same cooling protocol with the uninduced 32°C culture. While the cells are on ice, precool the centrifuge to 4°C and chill the necessary number of 35- to 50-ml plastic centrifuge tubes, labeled for induced and uninduced cells.
Make electrocompetent cells
- Transfer both the induced and uninduced cultures to the appropriately labeled chilled 35- to 50-ml centrifuge tubes. Centrifuge 7 min at 4600g (6700 rpm in a Sorvall SA-600 rotor), 4°C. Aspirate or pour off supernatant.
- Add 1 ml ice-cold distilled water to the cell pellet in the bottom of each tube and gently resuspend cells with a large pipet tip (do not vortex). Add another 30 ml ice-cold distilled water to each tube, seal, and gently invert to mix, again without vortexing. Centrifuge tubes again as in last step
- All subsequent resuspensions of cells should be done gently and without vortexing. Preparation of the cells for electroporation washes out any added chemical inducing agent.
- Decant the 30-ml supernatant very carefully from the soft pellet in each tube and resuspend each cell pellet in 1 ml ice-cold distilled water.
- Remove tubes from the centrifuge promptly. The pellet is very soft and care should be taken not to dislodge it, especially when processing multiple tubes.
- Transfer resuspended cells to microcentrifuge tubes. Microcentrifuge 30 to 60 sec at maximum speed, 4°C. Carefully aspirate supernatant. In each of the tubes, resuspend the cell pellet in 200 μl cold distilled water, which will provide enough material for four or five electroporations.
- Electrocompetent cells can be stored at −80°C after resuspending the cell pellet in 15% (v/v) glycerol. For highest efficiency, use freshly processed cells.
Introduce DNA by electroporation
- Chill the desired number of 0.1-cm electroporation cuvettes on ice. Turn on the electroporator and set to 1.80 kV.
- In microcentrifuge tubes on ice, 100 ng of single-stranded oligonucleotide with 50 to 100 μl of the suspension of induced or uninduced cells. Do the mixing and subsequent electroporation rapidly; do not leave the DNA-cell mixes on ice for extended periods. Be sure to include the following electroporation reactions and controls:
- Induced cells plus DNA.
- This is the culture that should yield the designed recombinants.
- Induced cells without DNA.
- This is a control to identify contamination, determine the reversion frequency, and obtain some idea of the efficiency of the selection.
- Uninduced cells plus DNA.
- This control tells whether there is some contaminating factor in the DNA that is contributing to the selected colonies.
- Induced cells plus DNA.
- Introduce the DNA into the cells by electroporation.
- The time constant should be greater than 5 msec for optimal results. Low time constants indicate problems with the cells, the DNA, or even the equipment.
- Immediately after electroporation, add 1 ml LB medium to the cuvette using a micropipettor with a 1000-μl pipet tip. Transfer the electroporation mix to sterile culture tubes and incubate the tubes with shaking at 30° to 34°C for 30min to 2 hr
Determine cell titers
- Make serial 1:10 dilutions of the electroporation mix through 10^−6 using M9medium or 1× TM buffer, dispensing 0.9 ml M9 or TM and 0.1 ml of the cell suspension per tube.
- The dilutions can be made in rich medium if a selection for antibiotic resistance is applied.
- To determine total viable cell count, spread 100 μl of 10^−5 and 10^−6 dilutions on LB plates (rich plates without drug). Incubate the plates at 30° to 34°C for 1 to 2 days.
- To determine recombinant cell count, plate cells on selective plates as follows depending on the anticipated recombinant frequency.
- If efficient recombination is expected, spread both 10 and 100 μl of the 10^−1 and 10^−2 dilutions.
- If low numbers of recombinants are expected, spread 100 μl each of a 1:5 and 1:10 dilution.
- For the no-DNA and uninduced controls, plate 200 μl directly on selective plates.
- Incubate plates at the appropriate temperature (30° to 34°C).
- At 30°C, colonies may take two days to come up on LB plates.
- Candidates should be purified by streaking for single colonies and retested for the appropriate phenotype.
Experiments and results
Design of the MAGE oligo
The design of the MAGE oligo is critical to the success of the procedure. Typically 90 bases long, with the first four 5’ bases phosphorothioated, the oligo must match the sequence of the region of interest (with the exception of the desired mutations) such that it will be incorporated into the lagging strand during replication. To determine which genomic strand to use as the template, it is necessary to determine whether the gene is in replichore 1 or 2 and whether it is on the + or - strand. The mismatches, insertions, and/or deletions in the sequence must be centered on the oligo, and there should be as few alterations as possible, since each change will lower the efficiency of incorporation into the genome.
Results
lacZ Replicore 1 G 365'242 -> A stop codon
*90mer oligo generated by optMAGE
GGCCGATACTGTCGTCGTCCCCTCAAACTGGCAGATGCACGGTTAAGATGCGCCCATCTACACCAACGTGACCTATCCCATTACGGTCAA
Testing of the system
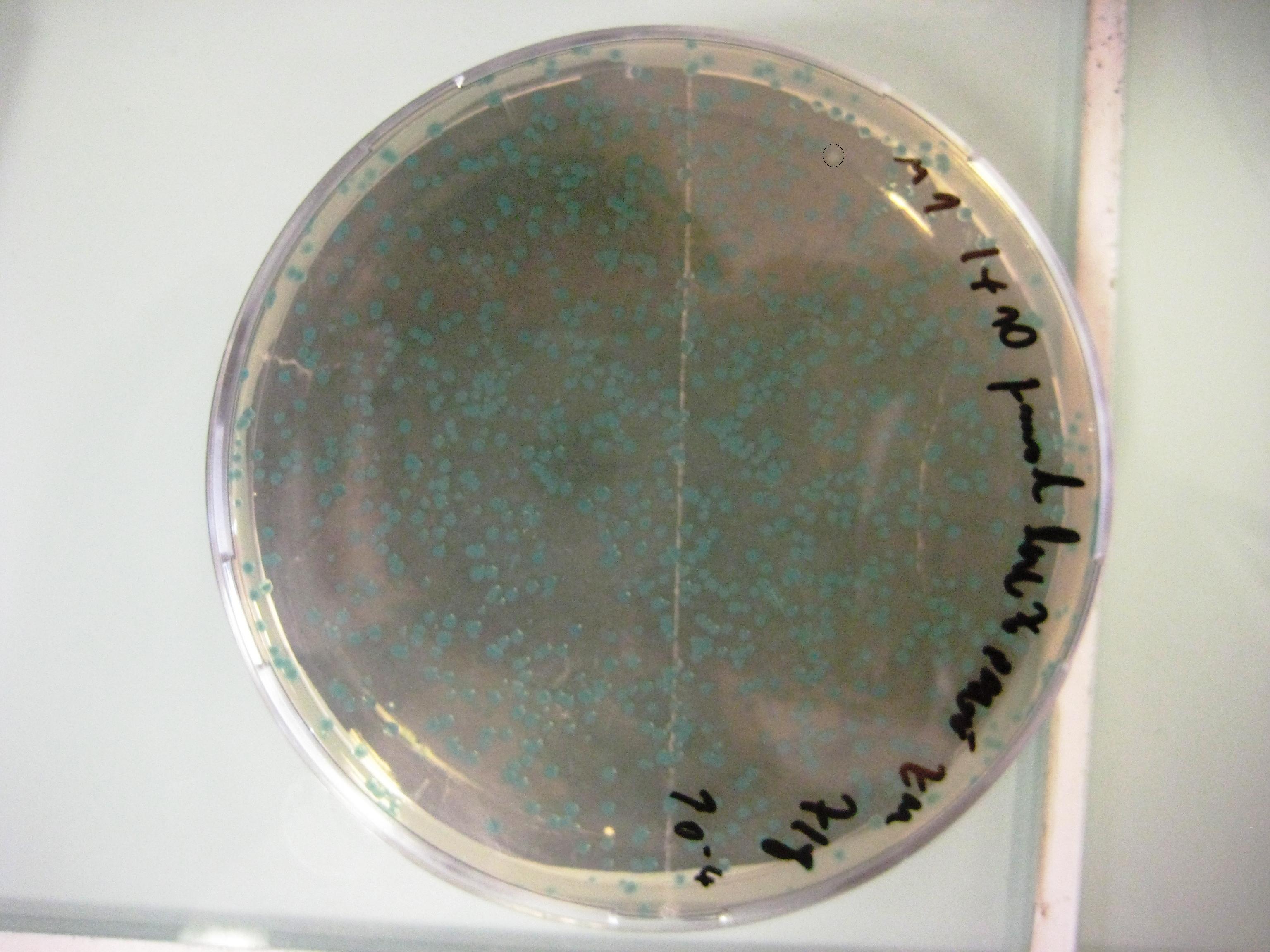
Unpurified Oligos
- Efficiency 0,0002 = 0,02% (33623 Blue Colonies / 7 White Colonies)
- no significant observation using different concentration of oligo
- no significant observation using phosphothiolated or normal oligo
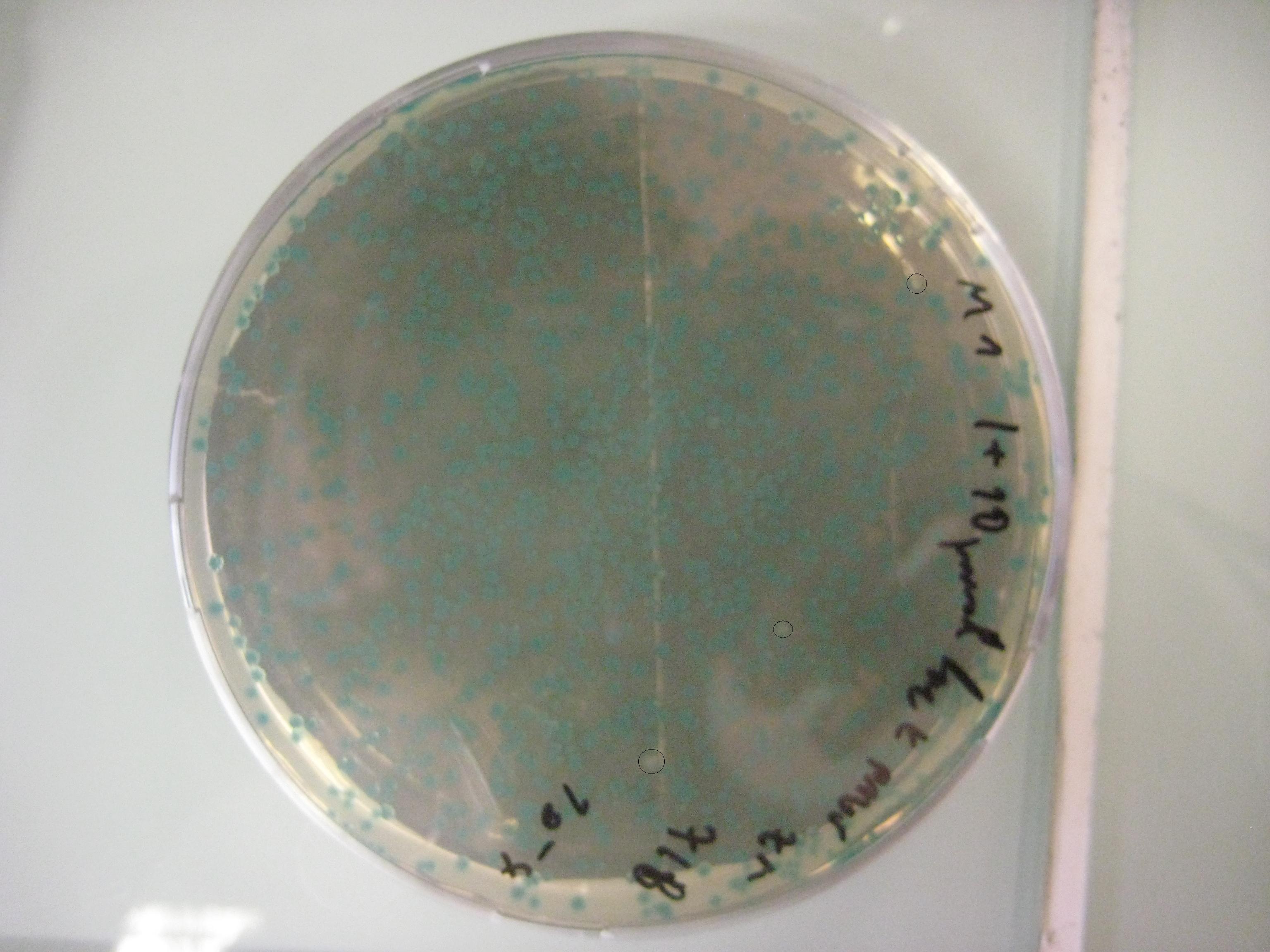
PAGE Purified Oligos
- Efficiency 0,0015 = 0,15% (4631 Blue Colonies / 7 White Colonies)
Discussion
As the "MAGE-by-hand" did not have the expected efficiency, we decided to stop with this part of the project.
References
- H. H. Wang et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894-898 (2009)
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment