Team:Bielefeld-Germany/Results/coli
From 2012.igem.org
| Line 3: | Line 3: | ||
<div id=page-title> | <div id=page-title> | ||
<span id=page-title-text> | <span id=page-title-text> | ||
| - | Laccase CueO from | + | Laccase CueO from <a href="https://www.atcc.org/ATCCAdvancedCatalogSearch/ProductDetails/tabid/452/Default.aspx?ATCCNum=BAA-1025&Template=bacteria"> <i>Escherichia coli</i> BL21 (DE3)</a> |
</span> | </span> | ||
</div> | </div> | ||
Revision as of 17:39, 26 September 2012
First some trials of shaking flask cultivations were made with changing parameters to identify the best conditions for the production of the laccase CueO from E. coli BL21 (DE3) named ECOL fused to a Histag. Because of no measured activity in the cell lysate a purification method was established (using Ni-NTA-Histag resin). The purified ECOL could be identified by SDS-PAGE (molecular weight of 53.4 kDa) as well as MALDI-TOF. The fractionated samples were also tested concerning their activity. A maximal activity of X was reached. After measuring activity of ECOL a scale up was made up to 3 L and then also up to 6 L.
Shaking Flask Cultivations
The first trials to produce ECOL were produced in shaking flask with various designs (from 100 mL-1 to 1 L flasks, with and without baffles) and under different conditions. The parameters we have changed during our screening experiments were temperature (27 °C,30 °C and 37 °C), concentrations of chloramphenicol (20-170 µg mL-1), various induction strategies (autoinduction and manual induction) and cultivation time (6 - 24 h). Further we cultivated with and without 0.25 mM CuCl2 to provide a sufficient amount of copper, which is needed for the active center of the laccase. Due to the screening experiments we identified the best conditions under which ECOL was expressed:
- flask design: shaking flask without baffles
- medium: autoinduction medium
- antibiotics: 60 µg mL-1 chloramphenicol
- temperature: 37 °C
- cultivation time: 12 h
The reproducibility and repeatability of the measured data and results were investigated for the shaking flask and bioreactor cultivation.
3 L Fermentation E. coli KRX with <partinfo>BBa_K863005</partinfo>
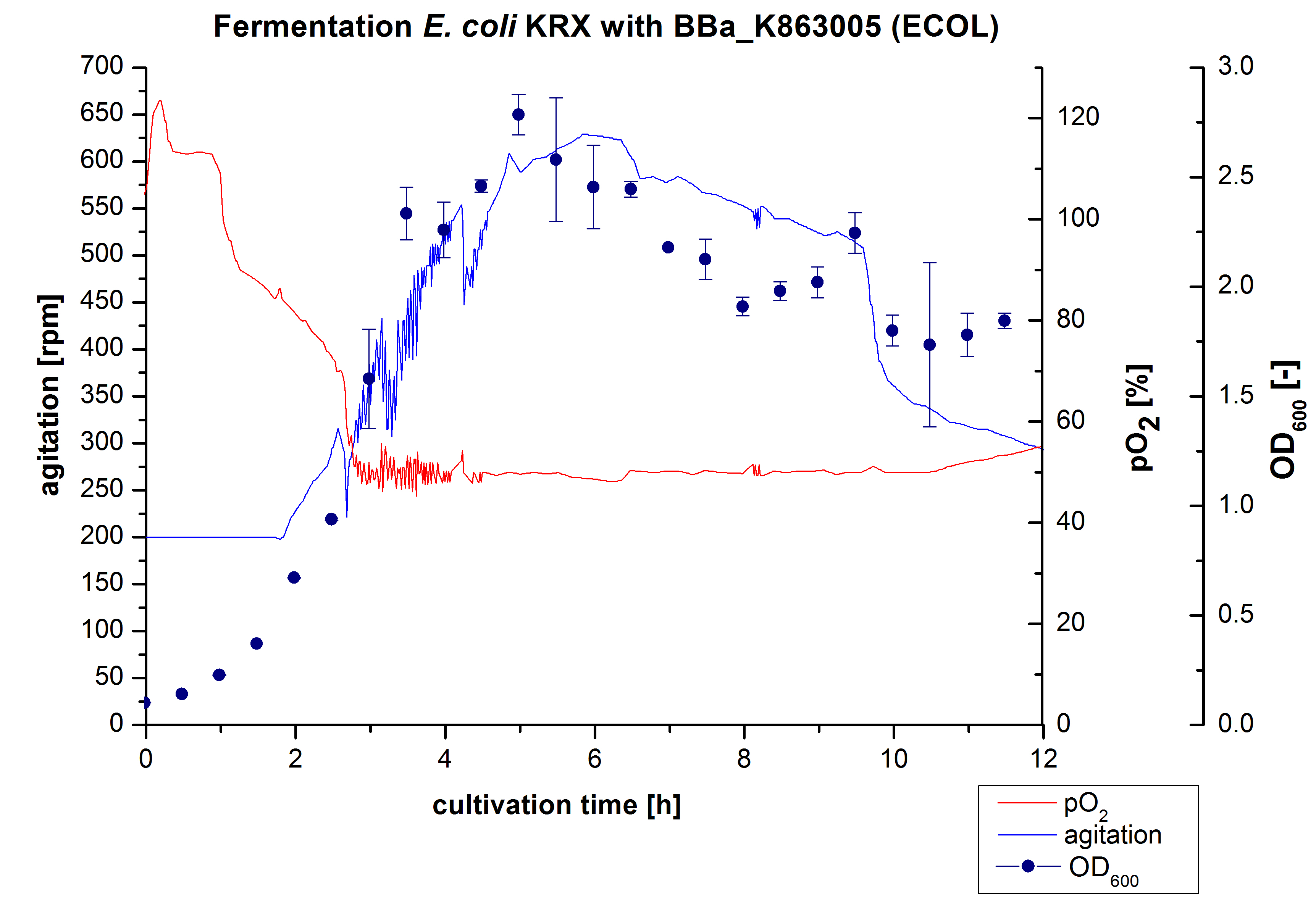
After the positive measurement of activity of ECOL we made a scale-up and fermented E. coli KRX with <partinfo>BBa_K863005</partinfo> in Infors Labfors with a total volume of 3 L. Agitation speed, pO2 and were determined and illustrated in figure 1. The exponential phase started after 1.5 hours of cultivation. The cell growth caused a decrease in pO2. After 2 hours of cultivation the agitation speed increased up to 629 rmp (5.9 hours) to hold the minimal pO2 level of 50 %. Then, after 4 hours there was a break in cell growth due to induction of protein expression. The maximal OD600 of 2.78 was reached after 5 hours. In comparison to E. coli KRX (OD600,max =4.86 after 8.5 hours) and to E. coli KRX with <partinfo>BBa_K863000</partinfo> (OD600,max =3.53 after 10 hours, time shift due to long lag phase) the OD600 max is lower. In the following hours, the OD600,max and the agitation speed decreased and the pO2 increased, which indicates the death phase of the cells. This is caused by the cell toxicity of ECOL (reference: [http://www.dbu.de/OPAC/ab/DBU-Abschlussbericht-AZ-13191.pdf DBU final report]). Therefore they were harvested after 12 hours.
Purification of ECOL
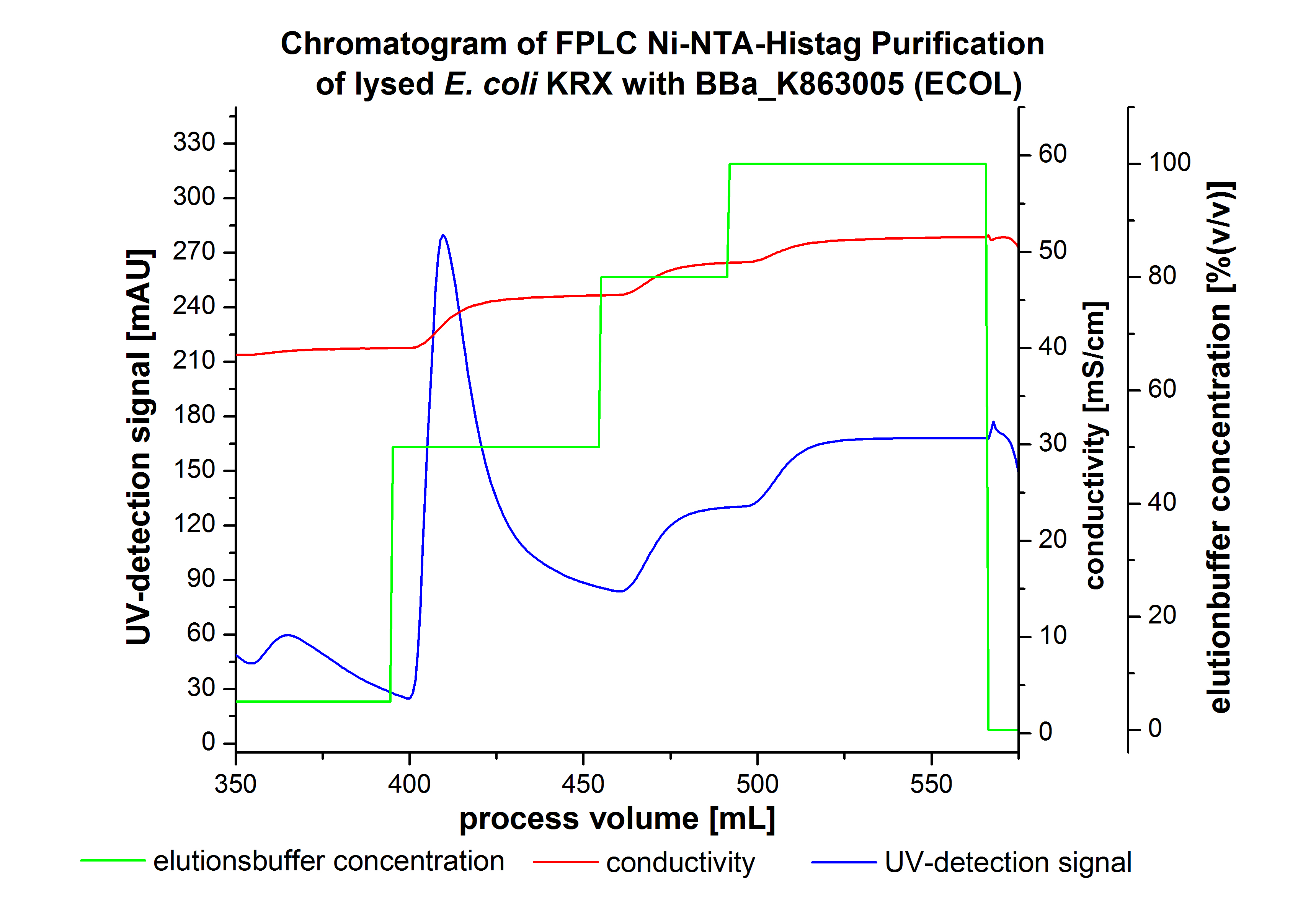
The harvested cells were resuspended in Ni-NTA-equilibration buffer, mechanically lysed by homogenization and centrifuged. The supernatant of the lysed cell paste was loaded on the Ni-NTA-column (15 mL Ni-NTA resin) with a flow rate of 1 mL min-1 cm-2. Then the column was washed by 10 column volumes (CV) Ni-NTA-equilibration buffer. The bound proteins were eluted by an increasing Ni-NTA-elution buffer step elution from 5 % (equates to 25 mM imidazol) with a length of 50 mL, to 50 % (equates to 250 mM imidazol) with a length of 60 mL, to 80 % (equates to 400 mM imidazol) with a length of 40 mL and finally to 100 % (equates to 500 mM imidazol) with a length of 80 mL. This strategies was chosen to improve the purification caused by a step by step increasing Ni-NTA-elution buffer concentration. The elution was collected in 10 mL fractions. Due to the high UV-detection signal of the loaded samples and to simplify the illustration of the detected product peak only the UV-detection signal of the wash step and the elution are shown. A typical chromatogram of purified laccases is illustrated here.The chromatogram of the ECOL-elution is shown in figure 2:
The chromatogram shows two distinguished peaks. The first peak was detected by a Ni-NTA-equilibration buffer concentration of 5 % (equates to 25 mM imidazol) and resulted from the elution of weakly bound proteins. After increasing the Ni-NTA-elutionbuffer concentration to 50 % (equates to 250 mM imidazol) a peak up to a UV-detection signal of 292 mAU was measured. The area of this peak indicates that a high amount of protein was eluted. The corresponding fractions were analyzed by SDS-PAGE to detect ECOL. There were no further peaks detectable. The following increasing UV-detection-signals equates to the imidazol concentration of the Ni-NTA-elution buffer. The corresponding SDS-PAGES are shown in figure 3.
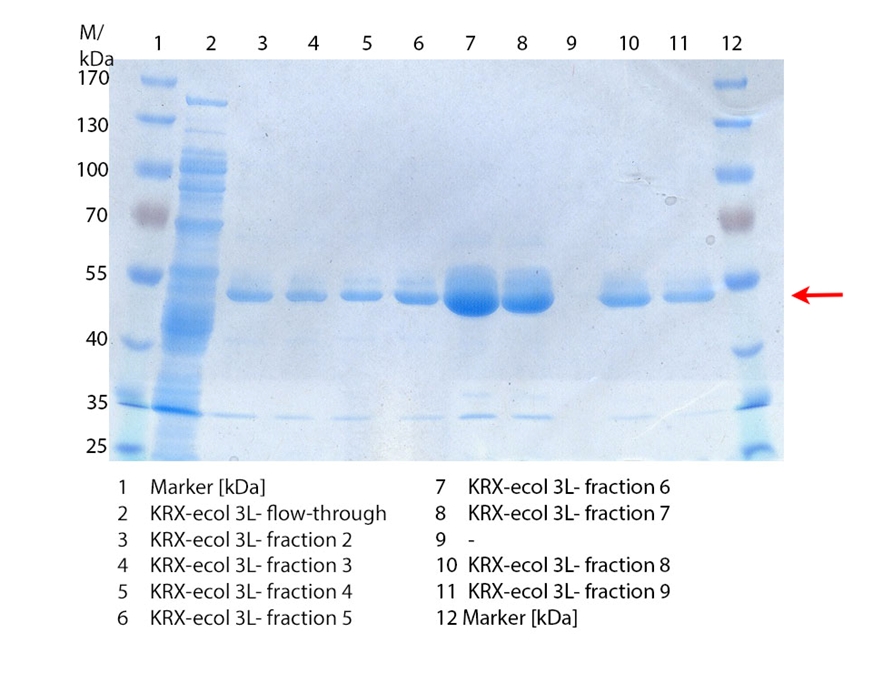
SDS-PAGE of ECOL purification
In figure 3 the SDS-PAGE of the Ni-NTA-His tag purification of the lysed culture (E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]) is illustrated including the flow-through and the fractions 2 to 9. The red arrow indicates the band of ECOL with a molecular weight of 53.4 kDa, which appears in all fractions. The strongest bands appear in fractions 6 and 7. These were the first two fractions (each 10 mL) eluted with 50 % Ni-NTA elution buffer (equates to 250 mM imidazol), in which the distinguished peak appeared. The second small band with a lower molecular weight was not identified.
Furthermore the bands were analyzed by MALDI-TOF and identified as CueO (ECOL).
6 L Fermentation E. coli KRX with <partinfo>BBa_K863005</partinfo>
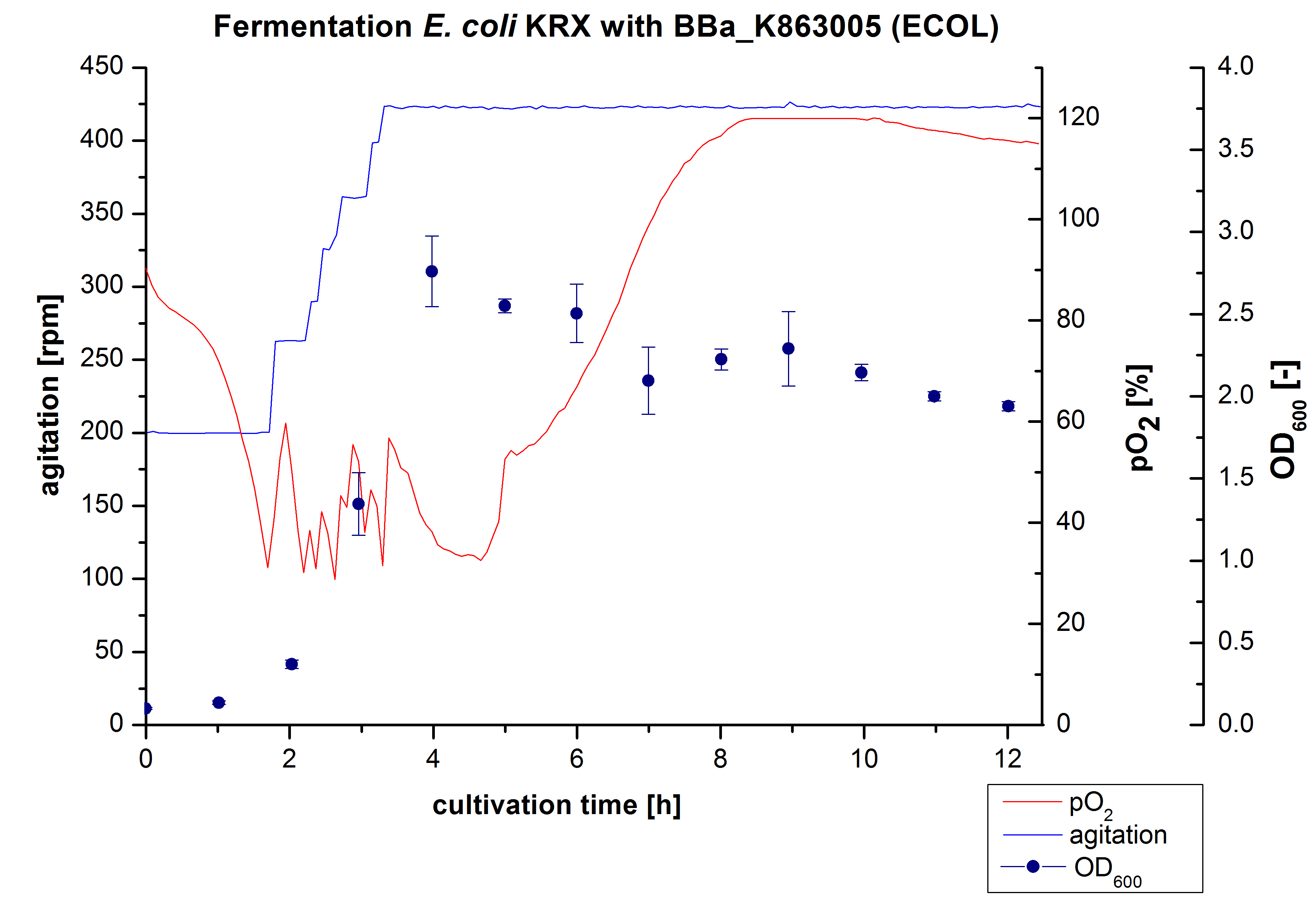
Another scale-up of the fermentation of E. coli KRX with <partinfo>BBa_K863005</partinfo> was made up to a final working volume of 6 L in Bioengineering NFL 22. Agitation speed, pO2 and OD600 were determined and illustrated in figure 3. There was no noticeable lag phase and the cells immediately began to grow. The cells were in an exponential phase between 2 and 4 hours of cultivation, which results in a decrease of pO2 value and therefore in an increase of agitation speed. After 4 hours of cultivation the maximal OD600 of 2.76 was reached, which is comparable to the 3 L fermentation of E. coli KRX with <partinfo>BBa_K863005</partinfo>. Due to induction of protein expression there is a break in cell growth. The death phase started, which is indicated by an increasing pO2 and a decreasing OD600. This demonstrates the cytotoxity of the laccases for E. coli, which was reported by the [http://www.dbu.de/OPAC/ab/DBU-Abschlussbericht-AZ-13191.pdf DBU]. In comparison to the fermentation of E. coli KRX with <partinfo>BBa_K863000</partinfo> under the same conditions (OD600,max= 3.53), the OD600,max was lower. Cells were harvested after 12 hours.
Purification of ECOL
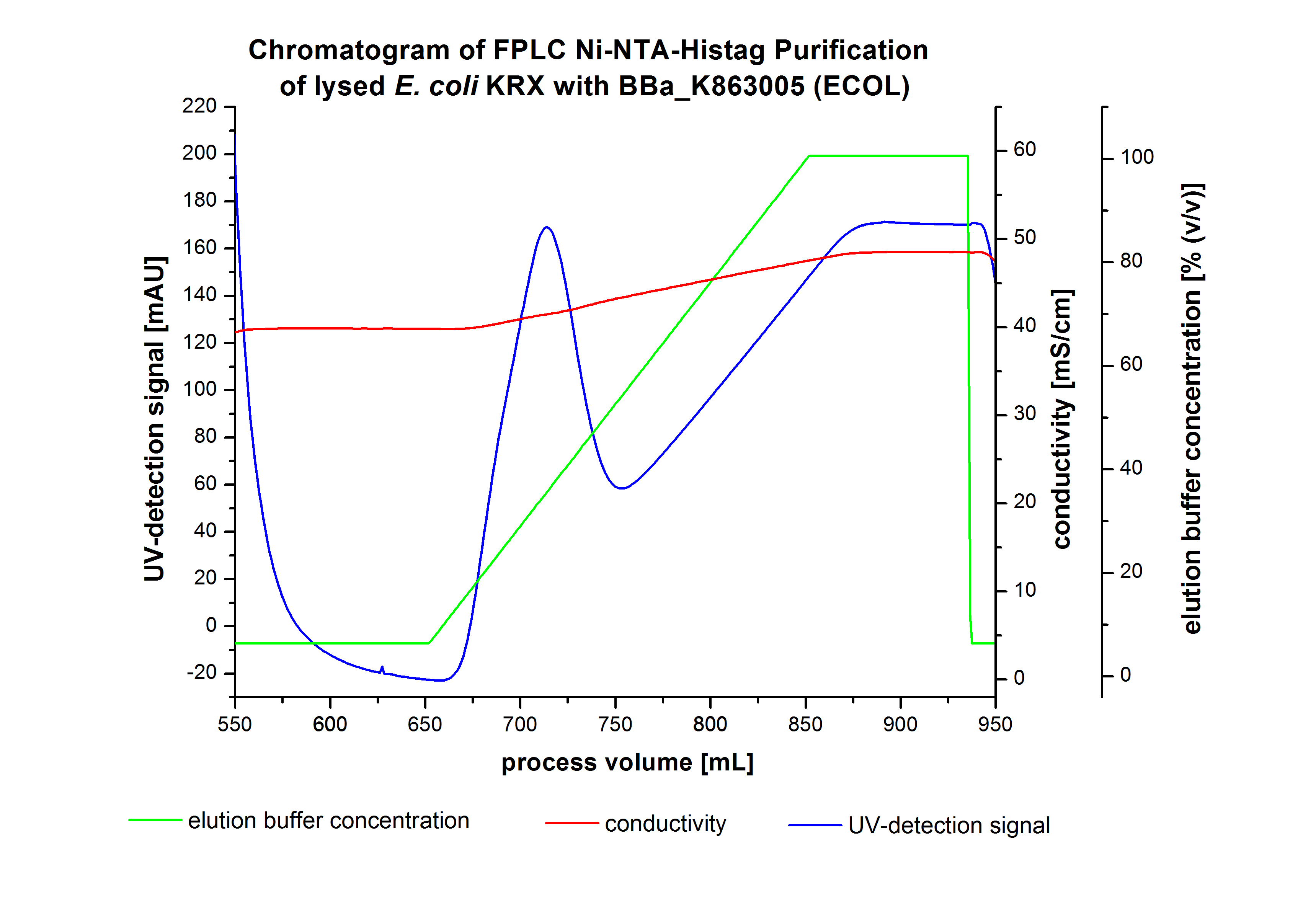
The harvested cells were resuspended in Ni-NTA-equilibratio nbuffer, mechanically lysed by homogenization and centrifuged. The supernatant of the lysed cell paste was loaded on the Ni-NTA-column (15 mL Ni-NTA resin) with a flow rate of 1 mL min-1 cm-2. The column was washed by 10 column volumes (CV) Ni-NTA-equilibration buffer. The bound proteins were eluted by an increasing Ni-NTA-elution buffer gradient from 0 % to 100 % with a length of 200 mL and the elution was collected in 10 mL fractions. Due to the high UV-detection signal of the loaded samples and to simplify the illustration of the detected product peak only the UV-detection signal of the wash step and the elution are shown. A typical chromatogram of purified laccases is illustrated here. The chromatogram of the ECOL elution is shown in figure 4:
After washing the column with 10 CV Ni-NTA-elution buffer the elution process was started. At a process volume of 670 mL to 750 mL the chromatogram shows a remarkable widespread peak (UV-detection signal 189 mAU) caused by the elution of a high amount of proteins. The run of the curve show a fronting. This can be explained by the elution of weakly bound proteins, which elutes at low imidazol concentrations. A better result could be achieved with a step elution strategy (see purification of the 3 L Fermentation above). To detect ECOL the corresponding fractions were analyzed by SDS-PAGE.
SDS-PAGES of ECOL purification
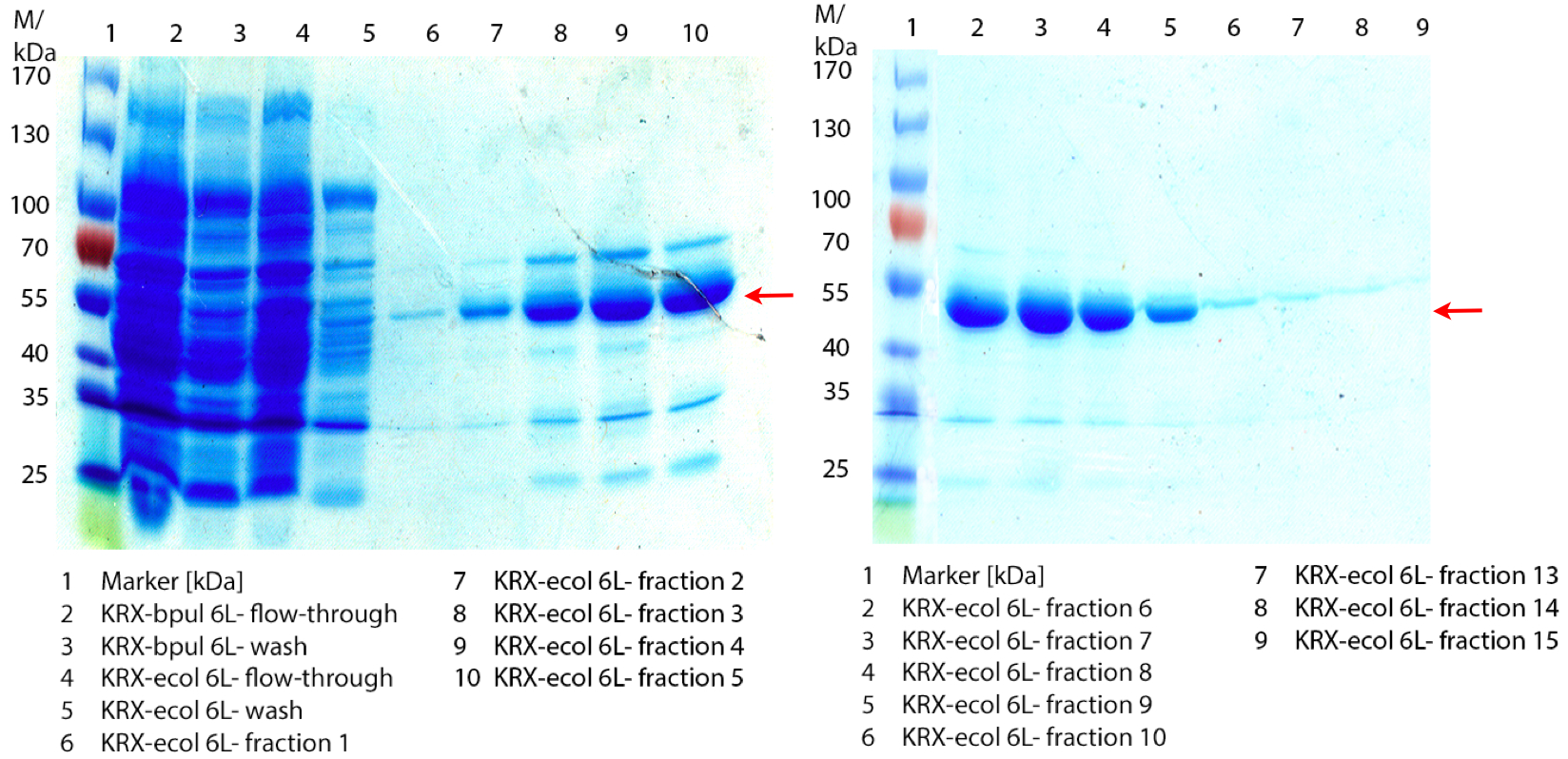
In figure 6 the result of the Ni-NTA-Histag purification of the lysed culture E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] (6 L fermentation) including the flow-through, wash and the fractions 1 to 15 (except from fraction 11/12) is shown. The red arrow indicates the band of ECOL with a molecular weight of 53.4 kDa, which appears in all fractions. The strongest bands appear from fractions 3 and 8 with a decreasing amount of other non-specific bands.
Furthermore the bands were analyzed by MALDI-TOF and identified as CueO (ECOL).
Activity Analysis of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 ECOL]
Initial activity tests of purified fractions
[http://partsregistry.org/Part:BBa_K863005 ECOL] pH optimum
[http://partsregistry.org/Part:BBa_K863005 ECOL] CuCl2 concentration
[http://partsregistry.org/Part:BBa_K863005 ECOL] activity at different temperatures
Impact of MeOH and acteonitrile on [http://partsregistry.org/Part:BBa_K863005 ECOL]
 "
"