Team:Bielefeld-Germany/Labjournal/week6
From 2012.igem.org
(Difference between revisions)
(→Monday June 4th) |
|||
| Line 1: | Line 1: | ||
{{Team:Bielefeld/Head}} | {{Team:Bielefeld/Head}} | ||
| + | |||
| + | |||
| + | <html> | ||
| + | <style type="text/css"> | ||
| + | |||
| + | ul {list-style-image:none;} | ||
| + | #bodyContent{ | ||
| + | background-color: white; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | |||
| + | <!-- navigator --> | ||
| + | <div id="nav" class="tabs"> | ||
| + | <div class="scroller"> | ||
| + | <ul style="list-style-type:none"> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/Starting"><strong>Prologue</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week1"><strong>Week 1</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week2"><strong>Week 2</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week3"><strong>Week 3</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week4"><strong>Week 4</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week5"><strong>Week 5</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week6"><strong>Week 6</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week7"><strong>Week 7</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week8"><strong>Week 8</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week9"><strong>Week 9</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week10"><strong>Week 10</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week11"><strong>Week 11</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week12"><strong>Week 12</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week13"><strong>Week 13</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week14"><strong>Week 14</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week15"><strong>Week 15</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week16"><strong>Week 16</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week17"><strong>Week 17</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week18"><strong>Week 18</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week19"><strong>Week 19</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week20"><strong>Week 20</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week21"><strong>Week 21</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week22"><strong>Week 22</strong></a></li> | ||
| + | |||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
Revision as of 10:07, 23 September 2012
Contents |
Week 6 (06/04 - 06/10/12)
Monday June 4th
- Team Cloning of Bacterial Laccases:
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
- PCR table
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
| Material | Volume |
|---|---|
| Buffer (10x Phusion) | 10µL |
| Phusion Polymerase | 0,5µL |
| dNTPs | 1µL |
| Primer Mix | 1µL |
| Template DNA | 1µL |
| DMSO | 1,5µL |
| Water | 35µL |
- PCR program
| Temperature | Time |
|---|---|
| 1) 98°C | 7 mins |
| 2) 98°C | 20 sec |
| 3) 55°C | 20 sec |
| 4) 72°C | 1 min |
| 5) 72°C | 3 min |
| 6) 12°C |
Cycle between step 2 and 4 35 times.
- Team Fungal and Plant Laccases:
- Primerdesign for isolating a laccase from Arababidopsis thaliana cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag.
Tuesday June 5th
- Team Student Academy:
- Transformation of a plasmid mixture of either pMTE cp46 His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] or pMTE cp46 His and [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] has a more intense fluorescence and was chosen for the experiment.
- Team Cloning of Bacterial Laccases:
- The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence.
- Again we did PCRs on T. thermophilus laccase and B. halodurans laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction.
- Team Activity Tests:
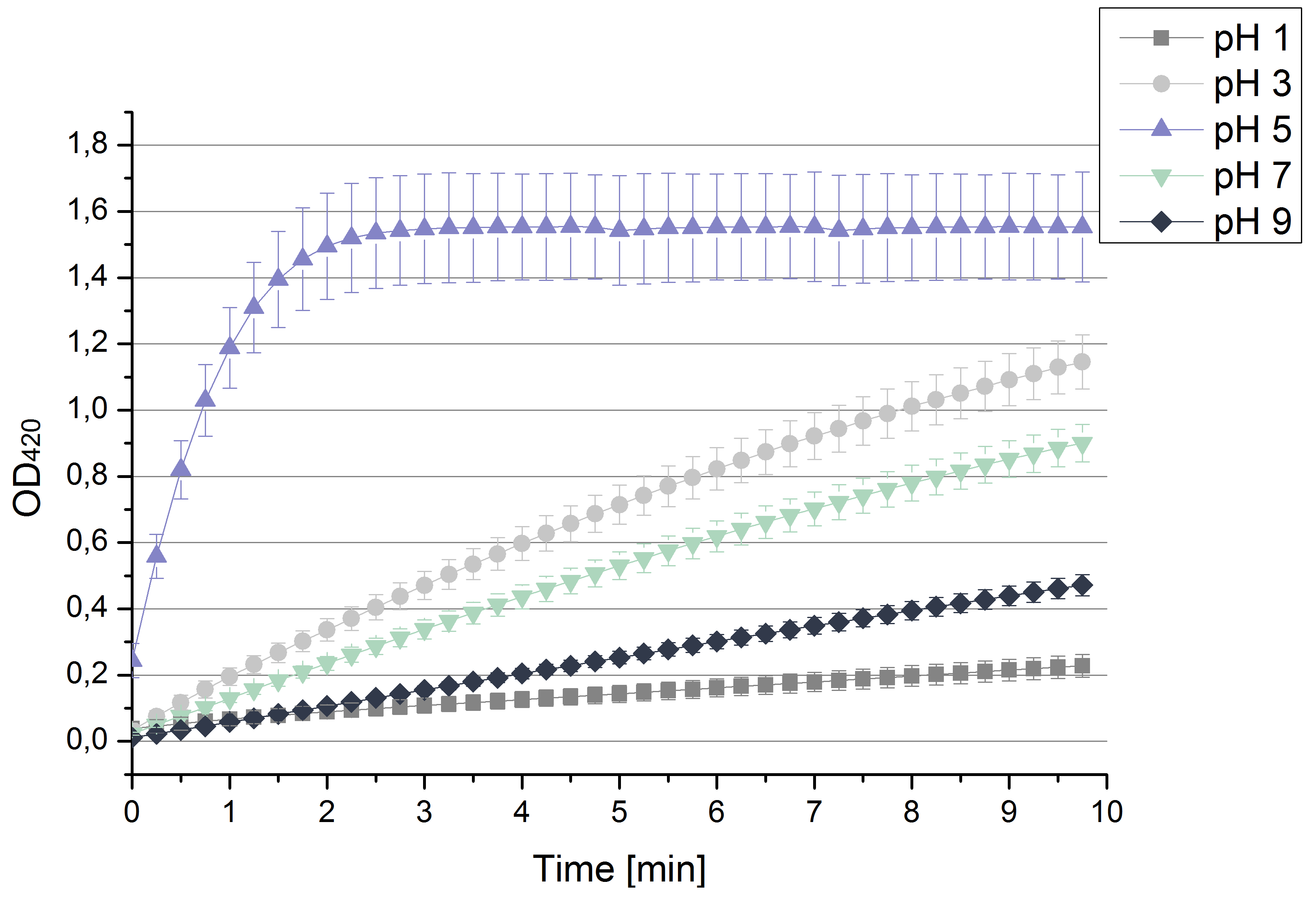
- After testing the T. versicolor laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned.
Wednesday June 6th
- Team Wiki: Yay for Team Wiki´s first entry. Our first steps with the iGEM Bielefeld 2012 Wiki contain thinking about contents, layouts, programming and responsibilities. Our first rules are:
- we are programming static pages in HTML and all the other pages (those that will be updated by all team members) in wiki code.
- we created all pages and will fill them up with some nice and beautiful content constantly from now on.
- Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon.
- Team Cloning of Bacterial Laccases: Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent E. coli KRX cells.
Thursday June 7th
- Team Student Academy:
- Repeating of Transformation of 06/05 to verify the function. It is reproducible :)
- Team Cloning of Bacterial Laccases: Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands.
- Team Modeling: becoming acquainted with matlab while reading the manual
Friday June 8th
- Team Cloning of Bacterial Laccases:
- We plated colonies for plasmid isolations on new plates and made a control restriction with NotI. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His.
Saturday June 9th
Sunday June 10th
| 55px | | | | | | | | | | |
 "
"