Team:Bielefeld-Germany/Labjournal/week5
From 2012.igem.org
(Difference between revisions)
(→Tuesday May 29th) |
(→Tuesday May 29th) |
||
| Line 16: | Line 16: | ||
*'''Team Bacterial Laccases:''' We made colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the ''E. coli'' laccase, the ''Xanthomonas campestris'' laccase and the ''B. pumilus'' laccase with the inducible t7 promotor and a HIS-tag. | *'''Team Bacterial Laccases:''' We made colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the ''E. coli'' laccase, the ''Xanthomonas campestris'' laccase and the ''B. pumilus'' laccase with the inducible t7 promotor and a HIS-tag. | ||
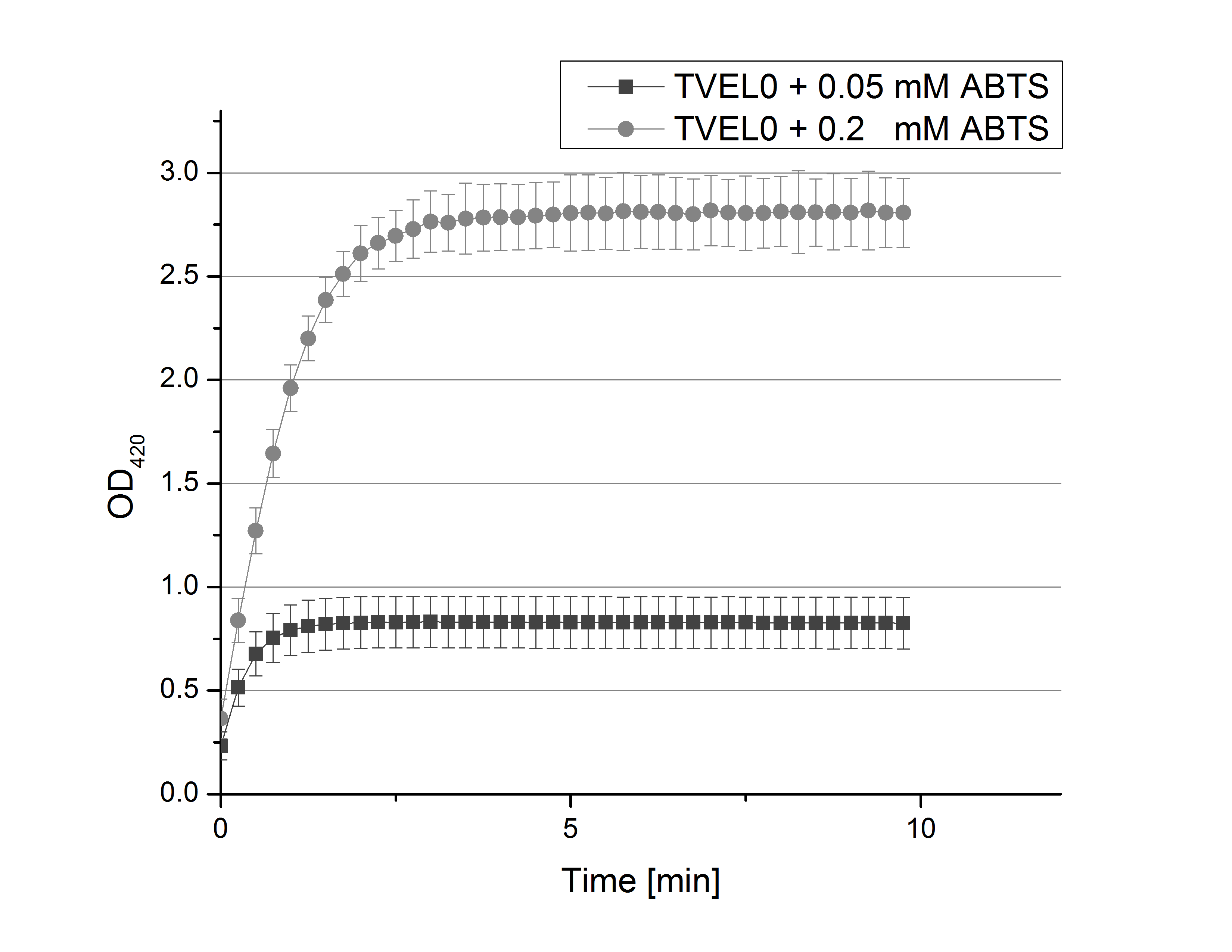
| - | [[ | + | [[File:Bielefeld2012_02_und_005_mM_ABTS.png|200px|thumb|right|Saturation curves of TVEL0 activity with 0.1 mM and 0.05 mM ABTS (n=24).]] |
*'''Team Activity Tests:''' | *'''Team Activity Tests:''' | ||
Revision as of 12:52, 19 September 2012
Contents |
Week 5 (05/28 - 06/03/12)
Monday May 28th
- Team bacterial laccase: Restriction of E. coli CueO, Xanthomonas campestris CopA and the B. pumilus laccase CotA and the pSB1C3 + RFP plasmid with the same restriction enzymes, ligation of the digested products and transformation in E.coli KRX cells.
- Since we have less CopA and Tth-laccase DNA, we set an PCR from the remaining PCR approach. For the reaction look PCR table
Tuesday May 29th
- Team Student Academy: E. coli Mach1 with pMTE cp46 His received from the working group “Fermentation Engineering”, University Bielefeld. Plasmid contains genes for GFP and kanamycine resistance. We plated it and made a liquid culture at 30°C. Result: There was an intense fluorescence. We made a glycerol stock and a plasmid isolation.
- Team Bacterial Laccases: We made colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the E. coli laccase, the Xanthomonas campestris laccase and the B. pumilus laccase with the inducible t7 promotor and a HIS-tag.
- Team Activity Tests:
Wednesday May 30th
- Team Bacterial Laccases: After plasmid isolation we cut our plasmids with NotI to see if the colony PCR was correct and our laccases are in the backbone. The agarose gel showed that for all of the different plasmids we had at least one plasmid with DNA band on the correct hight in agarose gel.
Thursday May 31st
- Team bacterial laccase: We sent the isolated pSB1C3 plasmids with inducible T7 promotor, the different laccase genes from Xanthomonas campestris CopA, B. pumilus CotA and E.coli CueO and HIS-Tag for sequencing.
Friday June 1st
- Team Activity Test: Today we prepared for our next measurements by setting up the sodium acetate buffer in different pHs. We choose to test the activity of TVEL0 in a pH-Gradient of 1,2,5,7 and 9.
Saturday June 2nd
Sunday June 3rd
- Team Bacterial Laccases:
- PCRs of genomic DNA from B. halodurans C-125 we ordered from DSMZ before. We handled the cells in the same way we did with T. thermphilus before and soluted the lyophlized cells in water and boiled them before PCR. After PCR we cleaned up the product. However the DNA amount was so low that we had to do the PCRs again.
| 55px | | | | | | | | | | |
 "
"