Team:TU Darmstadt/Labjournal/Transport
From 2012.igem.org
(Difference between revisions)
| Line 1: | Line 1: | ||
<html> | <html> | ||
| + | |||
<link rel="stylesheet" href="https://2012.igem.org/wiki/index.php?title=Team:TU_Darmstadt/css&action=raw&ctype=text/css" type="text/css" /> | <link rel="stylesheet" href="https://2012.igem.org/wiki/index.php?title=Team:TU_Darmstadt/css&action=raw&ctype=text/css" type="text/css" /> | ||
| + | |||
<div id="TUD"> | <div id="TUD"> | ||
| + | |||
<div id="off"> | <div id="off"> | ||
| + | |||
<div id="top"> | <div id="top"> | ||
| + | |||
<ul> | <ul> | ||
| + | |||
<li><a href="http://www.igem.tu-darmstadt.de/igem/projekt/index.en.jsp" title="iGEM">iGEM@TUD</a></li> | <li><a href="http://www.igem.tu-darmstadt.de/igem/projekt/index.en.jsp" title="iGEM">iGEM@TUD</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt" title="Home">Home</a></li> | <li><a href="/Team:TU_Darmstadt" title="Home">Home</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project" title="Project">Project</a><ul> | <li><a href="/Team:TU_Darmstadt/Project" title="Project">Project</a><ul> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project" title="Team:Overview">Overview</a></li> | <li><a href="/Team:TU_Darmstadt/Project" title="Team:Overview">Overview</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Degradation" title="Degradation">1. Degradation</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Degradation" title="Degradation">1. Degradation</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Transport" title="Transport">2. Transport</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Transport" title="Transport">2. Transport</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Metabolism" title="Metabolism">3. Metabolism</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Metabolism" title="Metabolism">3. Metabolism</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Material_Science" title="Material Science">4. Material Science</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Material_Science" title="Material Science">4. Material Science</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Simulation" title="Simulation">5. Simulation</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Simulation" title="Simulation">5. Simulation</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Ecology" title="Ecology">Ecology</a></li> | <li><a href="/Team:TU_Darmstadt/Project/Ecology" title="Ecology">Ecology</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Project/Philosophy" title="Philosophy">Philosophy</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Project/Philosophy" title="Philosophy">Philosophy</a></li></ul></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Team" title="Team:TU_Darmstadt/Team">Team</a><ul> | <li><a href="/Team:TU_Darmstadt/Team" title="Team:TU_Darmstadt/Team">Team</a><ul> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Team" title="Team:TU_Darmstadt/Team">Members</a></li> | <li><a href="/Team:TU_Darmstadt/Team" title="Team:TU_Darmstadt/Team">Members</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Supporters" title="Team:TU_Darmstadt/Supporters">Supporters</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Supporters" title="Team:TU_Darmstadt/Supporters">Supporters</a></li></ul></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Parts" title="BioBricks">BioBricks</a></li> | <li><a href="/Team:TU_Darmstadt/Parts" title="BioBricks">BioBricks</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Documentation" title="Documentation">Documentation</a><ul> | <li><a href="/Team:TU_Darmstadt/Documentation" title="Documentation">Documentation</a><ul> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Documentation" title="Documentation">Overview</a></li> | <li><a href="/Team:TU_Darmstadt/Documentation" title="Documentation">Overview</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Labjournal" title="Labjournal">Labjournal</a></li> | <li><a href="/Team:TU_Darmstadt/Labjournal" title="Labjournal">Labjournal</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Protocols" title="Protocols">Protocols</a></li> | <li><a href="/Team:TU_Darmstadt/Protocols" title="Protocols">Protocols</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Materials" title="Materials">Materials</a></li> | <li><a href="/Team:TU_Darmstadt/Materials" title="Materials">Materials</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Modeling" title="Modeling">Modeling</a></li> | <li><a href="/Team:TU_Darmstadt/Modeling" title="Modeling">Modeling</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Safety" title="Safety">Safety</a></li> | <li><a href="/Team:TU_Darmstadt/Safety" title="Safety">Safety</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Downloads" title="Downloads">Downloads</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Downloads" title="Downloads">Downloads</a></li></ul></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Human_Practice" title="Human Practice">Human Practice</a><ul> | <li><a href="/Team:TU_Darmstadt/Human_Practice" title="Human Practice">Human Practice</a><ul> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Human_Practice/Panel_Discussion " title="Panel Discussion">Panel Discussion</a></li> | <li><a href="/Team:TU_Darmstadt/Human_Practice/Panel_Discussion " title="Panel Discussion">Panel Discussion</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Human_Practice/Symposia" title="Symposia">Symposia</a></li> | <li><a href="/Team:TU_Darmstadt/Human_Practice/Symposia" title="Symposia">Symposia</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Human_Practice/Classes" title="Classes">Classes</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Human_Practice/Classes" title="Classes">Classes</a></li></ul></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Sponsors</a><ul> | <li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Sponsors</a><ul> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Overview</a></li> | <li><a href="/Team:TU_Darmstadt/Sponsors" title="Sponsors">Overview</a></li> | ||
| + | |||
<li><a href="/Team:TU_Darmstadt/Contact" title="Contact">Benefits</a></li></ul></li> | <li><a href="/Team:TU_Darmstadt/Contact" title="Contact">Benefits</a></li></ul></li> | ||
| + | |||
</ul> | </ul> | ||
| + | |||
<div id="igem"><a href="https://2012.igem.org/Main_Page"></a></div> | <div id="igem"><a href="https://2012.igem.org/Main_Page"></a></div> | ||
| + | |||
</div> | </div> | ||
| + | |||
<!-- end #menu --> | <!-- end #menu --> | ||
| + | |||
</html> | </html> | ||
| + | |||
<span style="font-size:188%;"><span style="color:#00689D;">Transport</span></span> | <span style="font-size:188%;"><span style="color:#00689D;">Transport</span></span> | ||
| + | |||
| + | |||
===Week 1 (21.-25.05.12)=== | ===Week 1 (21.-25.05.12)=== | ||
| + | |||
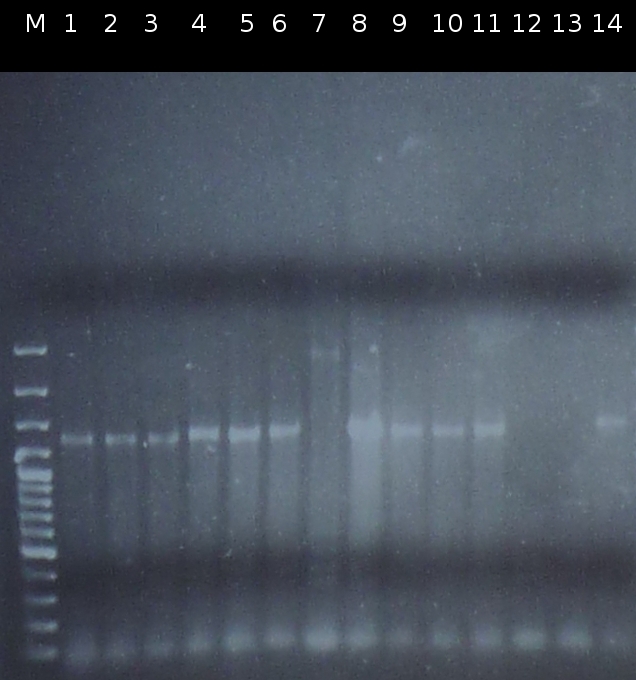
* First [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony PCR] of ''Comamonas testosteroni'' for isolation of the following genes: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5). (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). Analysis of [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * First [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony PCR] of ''Comamonas testosteroni'' for isolation of the following genes: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5). (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). Analysis of [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/1/15/W1_1_kolo_pcr.png | :https://static.igem.org/mediawiki/2012/1/15/W1_1_kolo_pcr.png | ||
| + | |||
* PCR product purification using the Promega-Kit | * PCR product purification using the Promega-Kit | ||
| + | |||
* Nanodrop measurements of the purified PCR products: | * Nanodrop measurements of the purified PCR products: | ||
| + | |||
:(1) : 19.5 ng/µl 260/280=1.58 | :(1) : 19.5 ng/µl 260/280=1.58 | ||
| + | |||
:(2) : 61.9 ng/µl 260/280=1.75 | :(2) : 61.9 ng/µl 260/280=1.75 | ||
| + | |||
:(3) : 75.1 ng/µl 260/280=1.75 | :(3) : 75.1 ng/µl 260/280=1.75 | ||
| + | |||
:(4) : 93.3 ng/µl 260/280=1.85 | :(4) : 93.3 ng/µl 260/280=1.85 | ||
| + | |||
:(5) : 91.3 ng/µl 260/280=1.58 | :(5) : 91.3 ng/µl 260/280=1.58 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of PCR products by SpeI and EcoRI, heat inactivation after digestion | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of PCR products by SpeI and EcoRI, heat inactivation after digestion | ||
| + | |||
* Analysis of restriction digest by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Analysis of restriction digest by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/1/15/w1_2_restriktionsverdau_s8.png | :https://static.igem.org/mediawiki/2012/1/15/w1_2_restriktionsverdau_s8.png | ||
| + | |||
* Purification of the bands gained form gel electrophoresis (Promega-Kit) | * Purification of the bands gained form gel electrophoresis (Promega-Kit) | ||
| + | |||
* Nanodrop measurements of the purified products: | * Nanodrop measurements of the purified products: | ||
| + | |||
:(1) : 8.3 ng/µl 260/280=1.94 | :(1) : 8.3 ng/µl 260/280=1.94 | ||
| + | |||
:(2) : 8.9 ng/µl 260/280=1.80 | :(2) : 8.9 ng/µl 260/280=1.80 | ||
| + | |||
:(3) : 13.0 ng/µl 260/280=1.57 | :(3) : 13.0 ng/µl 260/280=1.57 | ||
| + | |||
:(4) : 1.5 ng/µl 260/280=3.08 | :(4) : 1.5 ng/µl 260/280=3.08 | ||
| + | |||
:(5) : 3.7 ng/µl 260/280=2.26 | :(5) : 3.7 ng/µl 260/280=2.26 | ||
| + | |||
| + | |||
===week 2 (28.05.-01.06.12)=== | ===week 2 (28.05.-01.06.12)=== | ||
| + | |||
* Purified products from 1. week were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligated] into pSB1A2 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformed] into DH5a (in spite of low concentration) | * Purified products from 1. week were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligated] into pSB1A2 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformed] into DH5a (in spite of low concentration) | ||
| + | |||
* Overnight incubation on LB agar plates; no growth detectable | * Overnight incubation on LB agar plates; no growth detectable | ||
| + | |||
* Second approach to isolate the five genes from ''Comamonas t.'' using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * Second approach to isolate the five genes from ''Comamonas t.'' using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
* Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]; without results; modification of the PCR protocols: adding DMSO | * Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]; without results; modification of the PCR protocols: adding DMSO | ||
| + | |||
* Third approach to isolate the five genes from ''Comamonas t.'' by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * Third approach to isolate the five genes from ''Comamonas t.'' by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
* PCR product purification using the Promega-Kit: ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5)) | * PCR product purification using the Promega-Kit: ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5)) | ||
| + | |||
:https://static.igem.org/mediawiki/2012/f/fc/W2_1_kolo_pcr_2_s19.png | :https://static.igem.org/mediawiki/2012/f/fc/W2_1_kolo_pcr_2_s19.png | ||
| + | |||
:(1): no PCR-product | :(1): no PCR-product | ||
| + | |||
* Nanodrop measurements of the purified PCR products: | * Nanodrop measurements of the purified PCR products: | ||
| + | |||
:(2) : 56.1 ng/µl 260/280=1.87 | :(2) : 56.1 ng/µl 260/280=1.87 | ||
| + | |||
:(3) : 66.8 ng/µl 260/280=1.92 | :(3) : 66.8 ng/µl 260/280=1.92 | ||
| + | |||
:(4) : 72.8 ng/µl 260/280=1.87 | :(4) : 72.8 ng/µl 260/280=1.87 | ||
| + | |||
:(5) : 68.8 ng/µl 260/280=1.82 | :(5) : 68.8 ng/µl 260/280=1.82 | ||
| + | |||
| + | |||
===week 3 (04.-08.06.12)=== | ===week 3 (04.-08.06.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the purified PCR products and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (leftover of the 1. week) by SpeI and EcoRI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the purified PCR products and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (leftover of the 1. week) by SpeI and EcoRI | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 using SpeI and EcoRI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 using SpeI and EcoRI | ||
| + | |||
* Dephosphorylation of the restriction reactions by using antarctic Phosphatase | * Dephosphorylation of the restriction reactions by using antarctic Phosphatase | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the five genes into pSB1A2 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a (incubation at 37°C) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the five genes into pSB1A2 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a (incubation at 37°C) | ||
| + | |||
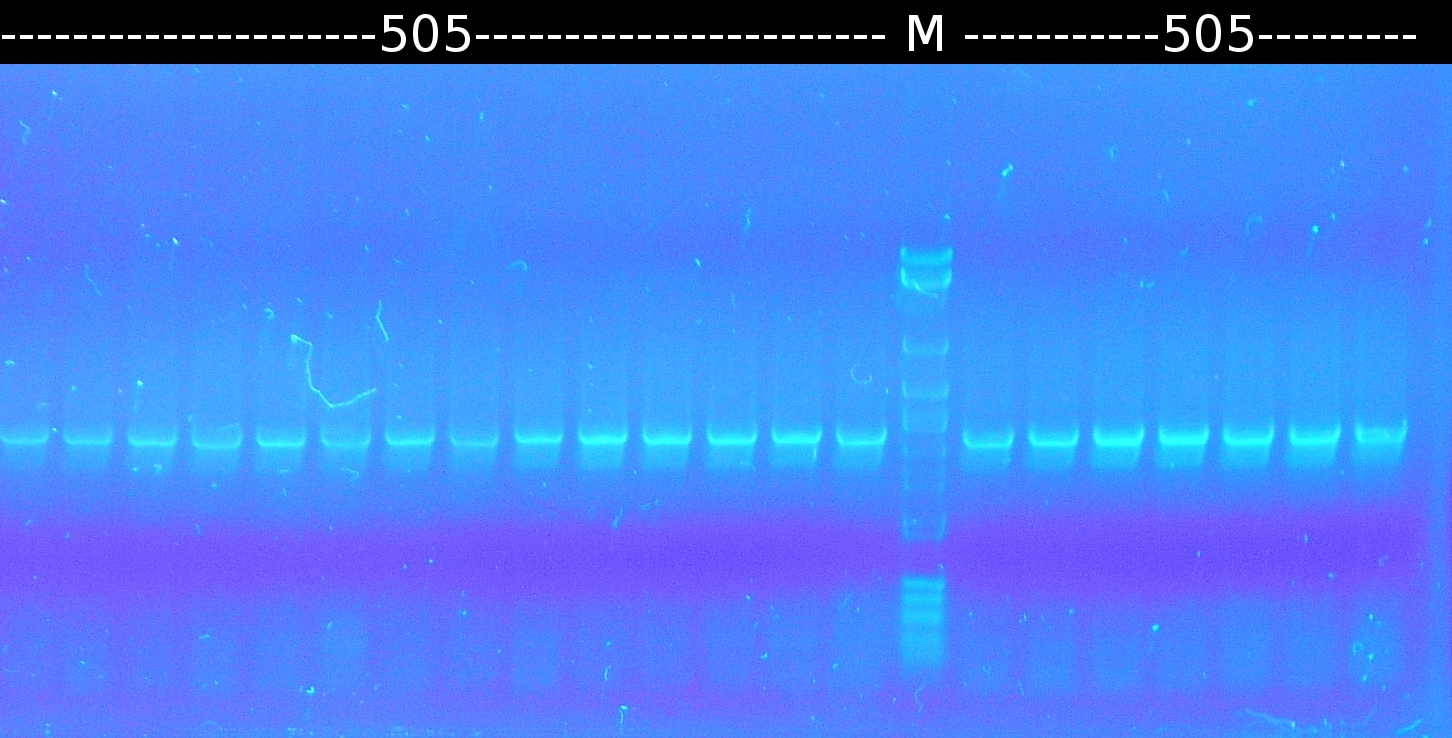
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches (Primer:[https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches (Primer:[https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]) | ||
| + | |||
* Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Only one of them showed the expected band. The band was excluded and purified via the Promega-Kit. | * Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Only one of them showed the expected band. The band was excluded and purified via the Promega-Kit. | ||
| + | |||
:https://static.igem.org/mediawiki/2012/1/11/W3_1_kolo_pcr_505_s23.png | :https://static.igem.org/mediawiki/2012/1/11/W3_1_kolo_pcr_505_s23.png | ||
| + | |||
* Nanodrop measurement of the purified product: | * Nanodrop measurement of the purified product: | ||
| + | |||
:(1.1) =8.9 ng/µl 260/280=2,22 | :(1.1) =8.9 ng/µl 260/280=2,22 | ||
| + | |||
:Because of low concentration and contamination the PCR was repeated and the product was purified again by the Promega-Kit. | :Because of low concentration and contamination the PCR was repeated and the product was purified again by the Promega-Kit. | ||
| + | |||
* Performing a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]-screen of the transformed cells using the Taq-Polymerase. Verification of 4 colonies per plate. (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * Performing a [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]-screen of the transformed cells using the Taq-Polymerase. Verification of 4 colonies per plate. (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
:PCR did not work | :PCR did not work | ||
| + | |||
* Testing the Taq-Polymerase for function. The Phu-Polymerase was used for comparison. Only the approach using the Phu-Polymerase showed the expected bands in [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. | * Testing the Taq-Polymerase for function. The Phu-Polymerase was used for comparison. Only the approach using the Phu-Polymerase showed the expected bands in [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. | ||
| + | |||
:https://static.igem.org/mediawiki/2012/5/5a/W3_2_taq-phu_s27.2.png | :https://static.igem.org/mediawiki/2012/5/5a/W3_2_taq-phu_s27.2.png | ||
| + | |||
* Screening was repeated with Phu-Polymerase; no bands visible | * Screening was repeated with Phu-Polymerase; no bands visible | ||
| + | |||
| + | |||
===week 4 (11.-15-06.12)=== | ===week 4 (11.-15-06.12)=== | ||
| + | |||
* Inoculation of LB-media with DH5a_pSB1A2 and DH5a_pSB1C3; overnight incubation at 37°C | * Inoculation of LB-media with DH5a_pSB1A2 and DH5a_pSB1C3; overnight incubation at 37°C | ||
| + | |||
* Miniprep of DH5a_pSB1A2 and DH5a_pSB1C3 using the Promega-Kit | * Miniprep of DH5a_pSB1A2 and DH5a_pSB1C3 using the Promega-Kit | ||
| + | |||
* Nanodrop measurements of the preparation: | * Nanodrop measurements of the preparation: | ||
| + | |||
:pSB1A2 : 136.1 ng/µl 260/280= 1.93 | :pSB1A2 : 136.1 ng/µl 260/280= 1.93 | ||
| + | |||
:pSB1C3 : 265.1 ng/µl 260/280= 1.89 | :pSB1C3 : 265.1 ng/µl 260/280= 1.89 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 and pSB1C3 with EcoRI and SpeI. Afterwards dephosphorylation by using Antarctic phosphatase. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 and pSB1C3 with EcoRI and SpeI. Afterwards dephosphorylation by using Antarctic phosphatase. | ||
| + | |||
* Analysis of the restriction products through [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]; for comparison the uncropped vectors were also analyzed. | * Analysis of the restriction products through [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]; for comparison the uncropped vectors were also analyzed. | ||
| + | |||
:https://static.igem.org/mediawiki/2012/6/69/W4_1_restriktion_der_vektoren.png | :https://static.igem.org/mediawiki/2012/6/69/W4_1_restriktion_der_vektoren.png | ||
| + | |||
:pSB1C3 shows several bands (Insert, vector without insert, linearized vector with insert); concentration of pSB1A2 is very low, only one band visible | :pSB1C3 shows several bands (Insert, vector without insert, linearized vector with insert); concentration of pSB1A2 is very low, only one band visible | ||
| + | |||
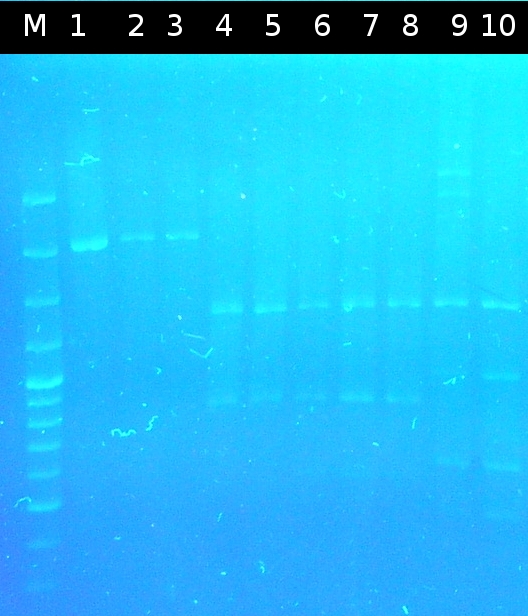
* Approach to isolate the following genes from ''Comamonas t.'' by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * Approach to isolate the following genes from ''Comamonas t.'' by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
* Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Analysis of PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/9/9f/W4_2_koloPCRcoma_197-503-162.png | :https://static.igem.org/mediawiki/2012/9/9f/W4_2_koloPCRcoma_197-503-162.png | ||
| + | |||
:Isolation of (1) and (5) did not work | :Isolation of (1) and (5) did not work | ||
| + | |||
* PCR product purification by using the Promega-Kit | * PCR product purification by using the Promega-Kit | ||
| + | |||
* Nanodrop measurements of the purified PCR products: | * Nanodrop measurements of the purified PCR products: | ||
| + | |||
:(2) : 18.3 ng/µl 260/280=2.03 | :(2) : 18.3 ng/µl 260/280=2.03 | ||
| + | |||
:(3) : 21.2 ng/µl 260/280=1.95 | :(3) : 21.2 ng/µl 260/280=1.95 | ||
| + | |||
:(4) : 23.9 ng/µl 260/280=2.05 | :(4) : 23.9 ng/µl 260/280=2.05 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of purified PCR products (2), (3) and (4) using SpeI and EcoRI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of purified PCR products (2), (3) and (4) using SpeI and EcoRI | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the digested genes ((2), (3), (4)) into pSB1A2 and pSB1C3 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the digested genes ((2), (3), (4)) into pSB1A2 and pSB1C3 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of DH5a with pSB1A2_197, pSB1A2_503, pSB1A2_162, pSB1C3_197, pSB1C3_503, pSB1C3_162 (6 different transformation approaches). Overnight incubation at 37°C | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of DH5a with pSB1A2_197, pSB1A2_503, pSB1A2_162, pSB1C3_197, pSB1C3_503, pSB1C3_162 (6 different transformation approaches). Overnight incubation at 37°C | ||
| + | |||
* Screening of the transformants by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). Verification of 6 colonies for every transformation. Afterwards the PCR products were analyzed by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Screening of the transformants by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). Verification of 6 colonies for every transformation. Afterwards the PCR products were analyzed by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/d/dd/W4_3_koloPCRscreen1.png | :https://static.igem.org/mediawiki/2012/d/dd/W4_3_koloPCRscreen1.png | ||
| + | |||
:https://static.igem.org/mediawiki/2012/4/40/W4_4_koloPCRscreen2.png | :https://static.igem.org/mediawiki/2012/4/40/W4_4_koloPCRscreen2.png | ||
| + | |||
:positive colonies are marked | :positive colonies are marked | ||
| + | |||
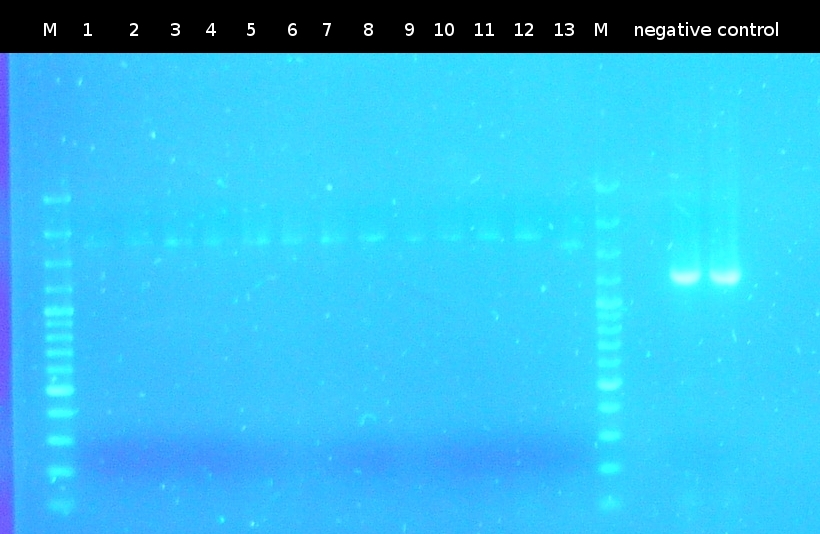
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR] of ''Comamonas t.'' to isolate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) und [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR] of ''Comamonas t.'' to isolate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) und [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
* Analysis of PCR products (Colony-PCR) and the ligation approach by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Analysis of PCR products (Colony-PCR) and the ligation approach by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/f/f7/W4_5_ligation%2BkoloPCR505-322.png | :https://static.igem.org/mediawiki/2012/f/f7/W4_5_ligation%2BkoloPCR505-322.png | ||
| + | |||
:PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] shows only the band of the undesired smaller product | :PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] shows only the band of the undesired smaller product | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] into pSB1A2 and pSB1C3 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] into pSB1A2 and pSB1C3 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of DH5a with pSB1A2_322 und pSB1C3_322. [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) used to check for successful transformation<!-- bild fehlt!! --> | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of DH5a with pSB1A2_322 und pSB1C3_322. [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) used to check for successful transformation<!-- bild fehlt!! --> | ||
| + | |||
* Isolation of ''Comamonas t.'' genome, because it seems impossible to amplificate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) from a fluid ''Comamonas t.'' culture. | * Isolation of ''Comamonas t.'' genome, because it seems impossible to amplificate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) from a fluid ''Comamonas t.'' culture. | ||
| + | |||
| + | |||
===week 5 (18.-22.06.12)=== | ===week 5 (18.-22.06.12)=== | ||
| + | |||
* Introduction of a nomenclature: A/C gene/colony: pSB1A2 = A; pSB1C3 = C; gene: tctA 505aa = 1, tctB 197aa = 2, tctA 503aa = 3, tctB 162aa = 4, tphC 322aa = 5; colony = 1...X | * Introduction of a nomenclature: A/C gene/colony: pSB1A2 = A; pSB1C3 = C; gene: tctA 505aa = 1, tctB 197aa = 2, tctA 503aa = 3, tctB 162aa = 4, tphC 322aa = 5; colony = 1...X | ||
| + | |||
* Producing fluid cultures of the positive transformants | * Producing fluid cultures of the positive transformants | ||
| + | |||
* Miniprep of the fluid culures by using the Frementas-Kit. Afterwards Nanodrop measurements of the preparations | * Miniprep of the fluid culures by using the Frementas-Kit. Afterwards Nanodrop measurements of the preparations | ||
| + | |||
:https://static.igem.org/mediawiki/2012/8/8f/W5_1_positive_transformanden.png | :https://static.igem.org/mediawiki/2012/8/8f/W5_1_positive_transformanden.png | ||
| + | |||
:A2.3 : 125.9 ng/µl 260/280= 1.62 | :A2.3 : 125.9 ng/µl 260/280= 1.62 | ||
| + | |||
:A3.1 : 103.3 ng/µl 260/280= 1.72 | :A3.1 : 103.3 ng/µl 260/280= 1.72 | ||
| + | |||
:A4.6 : 147.2 ng/µl 260/280= 1.70 | :A4.6 : 147.2 ng/µl 260/280= 1.70 | ||
| + | |||
:A4.5 : 108.7 ng/µl 260/280= 1.86 | :A4.5 : 108.7 ng/µl 260/280= 1.86 | ||
| + | |||
:A5.12 : 88.0 ng/µl 260/280= 1.87 | :A5.12 : 88.0 ng/µl 260/280= 1.87 | ||
| + | |||
:A5.8 : 85.01 ng/µl 260/280= 1.78 | :A5.8 : 85.01 ng/µl 260/280= 1.78 | ||
| + | |||
:A3.2 : 87.3 ng/µl 260/280= 1.81 | :A3.2 : 87.3 ng/µl 260/280= 1.81 | ||
| + | |||
:C4.6 : 70.5 ng/µl 260/280= 1.81 | :C4.6 : 70.5 ng/µl 260/280= 1.81 | ||
| + | |||
:C4.1 : 41.2 ng/µl 260/280= 1.85 | :C4.1 : 41.2 ng/µl 260/280= 1.85 | ||
| + | |||
:C4.4 : 104.2 ng/µl 260/280= 1.85 | :C4.4 : 104.2 ng/µl 260/280= 1.85 | ||
| + | |||
:C2.1 : 132.2 ng/µl 260/280= 1.85 | :C2.1 : 132.2 ng/µl 260/280= 1.85 | ||
| + | |||
:C2.6 : 188.1 ng/µl 260/280= 1.75 | :C2.6 : 188.1 ng/µl 260/280= 1.75 | ||
| + | |||
* [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] using a rest of purified [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]), subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] using a rest of purified [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]), subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
* Another [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] using the transformants (2)-(5) as template (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]); analyzing the PCR product by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Another [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] using the transformants (2)-(5) as template (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]); analyzing the PCR product by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/1/15/W5_2_screen.png | :https://static.igem.org/mediawiki/2012/1/15/W5_2_screen.png | ||
| + | |||
:3 positive transformants were picked for inoculation of fluid media (A34, A43; C513) | :3 positive transformants were picked for inoculation of fluid media (A34, A43; C513) | ||
| + | |||
* Miniprep (Fermentas Kit) of the fluid culures and Nanodrop measurements: | * Miniprep (Fermentas Kit) of the fluid culures and Nanodrop measurements: | ||
| + | |||
:A2.4 : 202,1 ng/µl 260/280= 1,63 | :A2.4 : 202,1 ng/µl 260/280= 1,63 | ||
| + | |||
:A2.5 : 126,5 ng/µl 260/280= 1,90 | :A2.5 : 126,5 ng/µl 260/280= 1,90 | ||
| + | |||
:A2.6 : 107,5 ng/µl 260/280= 1,81 | :A2.6 : 107,5 ng/µl 260/280= 1,81 | ||
| + | |||
:A4.2 : 133,8 ng/µl 260/280= 1,59 | :A4.2 : 133,8 ng/µl 260/280= 1,59 | ||
| + | |||
:C2.3 : 187,8 ng/µl 260/280= 1,87 | :C2.3 : 187,8 ng/µl 260/280= 1,87 | ||
| + | |||
:C2.4 : 270,7 ng/µl 260/280= 1,80 | :C2.4 : 270,7 ng/µl 260/280= 1,80 | ||
| + | |||
:C4.1 : 103,8 ng/µl 260/280= 1,80 | :C4.1 : 103,8 ng/µl 260/280= 1,80 | ||
| + | |||
* The Miniprep products were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digested] by restriction enzymes, each was cut once with EcoRI and twice with EcoRI/SpeI. Analisys of the digested products by [[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * The Miniprep products were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digested] by restriction enzymes, each was cut once with EcoRI and twice with EcoRI/SpeI. Analisys of the digested products by [[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/4/4b/W5_3_miniprep1x2x.png | :https://static.igem.org/mediawiki/2012/4/4b/W5_3_miniprep1x2x.png | ||
| + | |||
| + | |||
===week 6 (25.-29.06.12)=== | ===week 6 (25.-29.06.12)=== | ||
| + | |||
* Further [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of ''Comamonas t.'' (3 approaches) and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic ''Comamonas t.'' DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches each (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]). Subsequent analisys of the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. All PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] show only the band of the undesired smaller product. | * Further [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of ''Comamonas t.'' (3 approaches) and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic ''Comamonas t.'' DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches each (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]). Subsequent analisys of the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. All PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] show only the band of the undesired smaller product. | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] off the digested genes into pSB1A2 and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. Overnight incubation at 37°C | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] off the digested genes into pSB1A2 and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. Overnight incubation at 37°C | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR] of ''Comamonas t.'' and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic ''Comamonas t.'' DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1). [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) used as control (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR] of ''Comamonas t.'' and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic ''Comamonas t.'' DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1). [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) used as control (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) | ||
| + | |||
:https://static.igem.org/mediawiki/2012/c/ce/W6_1_PCR_505.png | :https://static.igem.org/mediawiki/2012/c/ce/W6_1_PCR_505.png | ||
| + | |||
* Miniprep and Nanodrop measurement of: | * Miniprep and Nanodrop measurement of: | ||
| + | |||
:A3.1 : 111.6 ng/µl 260/280= 1.72 | :A3.1 : 111.6 ng/µl 260/280= 1.72 | ||
| + | |||
:A3.4 : 79.5 ng/µl 260/280= 1.66 | :A3.4 : 79.5 ng/µl 260/280= 1.66 | ||
| + | |||
:C4.6 : 69.6 ng/µl 260/280= 1.76 | :C4.6 : 69.6 ng/µl 260/280= 1.76 | ||
| + | |||
:A4.2 : 96.9 ng/µl 260/280= 1.62 | :A4.2 : 96.9 ng/µl 260/280= 1.62 | ||
| + | |||
:A2.4 : 109.7 ng/µl 260/280= 1.72 | :A2.4 : 109.7 ng/µl 260/280= 1.72 | ||
| + | |||
:A2.5 : 160.8 ng/µl 260/280= 1.79 | :A2.5 : 160.8 ng/µl 260/280= 1.79 | ||
| + | |||
:A5.13 : 188.7 ng/µl 260/280= 1.85 | :A5.13 : 188.7 ng/µl 260/280= 1.85 | ||
| + | |||
:A5.12 : 114.5 ng/µl 260/280= 1.81 | :A5.12 : 114.5 ng/µl 260/280= 1.81 | ||
| + | |||
* Checking the Miniprep with [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * Checking the Miniprep with [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:https://static.igem.org/mediawiki/2012/7/7c/W6_2_miniprep.png | :https://static.igem.org/mediawiki/2012/7/7c/W6_2_miniprep.png | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR]-Screen of the transformants (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]) and analisys of the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR Colony-PCR]-Screen of the transformants (Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]) and analisys of the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] | ||
| + | |||
:No bands visible | :No bands visible | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] into pSB1C3 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. No colonies | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] into pSB1C3 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. No colonies | ||
| + | |||
* The following vectors were sequenced: pSB1A2_197.4, pSB1A2_197.5, psSB1A2_503.1, pSB1A2_503.4, pSB1A2_162.2, pSB1C3_162.6, pSB1A2_322.12, pSB1A2_322.13 | * The following vectors were sequenced: pSB1A2_197.4, pSB1A2_197.5, psSB1A2_503.1, pSB1A2_503.4, pSB1A2_162.2, pSB1C3_162.6, pSB1A2_322.12, pSB1A2_322.13 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] using EcoRI/SpeI. [[Ligation]] into pSB1C3 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] using EcoRI/SpeI. [[Ligation]] into pSB1C3 and subsequently [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. | ||
| + | |||
* Preperations for pure culture: [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of A2.4, A2.5, A3.1, A3.4, A4.2, C4.6, A5.12, A5.13 (2 colonies each, Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) and afterwards checking the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Positive colonies were picked, streaked onto fresh media and incubated again. | * Preperations for pure culture: [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of A2.4, A2.5, A3.1, A3.4, A4.2, C4.6, A5.12, A5.13 (2 colonies each, Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB197 tctB_197 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]) and afterwards checking the PCR products by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Positive colonies were picked, streaked onto fresh media and incubated again. | ||
| + | |||
* As A5.12 and A5.13 are missing the right insert the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] was repeated (5 and 6 colonies were used, Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). To provide additional monitoring the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] was repeated for A2.5_2, A3.1_2 und A4.2_2. 2 positive colonies were again streaked onto fresh media. | * As A5.12 and A5.13 are missing the right insert the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] was repeated (5 and 6 colonies were used, Primer: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). To provide additional monitoring the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] was repeated for A2.5_2, A3.1_2 und A4.2_2. 2 positive colonies were again streaked onto fresh media. | ||
| + | |||
* The Arabinose-Promotor ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara]) was isolated from pbad_turbo_gfp and colonies containing this plasmid (3 approaches each). Approach T2 is further in use. | * The Arabinose-Promotor ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara]) was isolated from pbad_turbo_gfp and colonies containing this plasmid (3 approaches each). Approach T2 is further in use. | ||
| + | |||
:https://static.igem.org/mediawiki/2012/f/fd/W6_3_ara.png | :https://static.igem.org/mediawiki/2012/f/fd/W6_3_ara.png | ||
| + | |||
| + | |||
===week 7 (02.-06.07.12)=== | ===week 7 (02.-06.07.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] and pXylE using EcoRI and SpeI. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the restricted products and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. No colonies | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] and pXylE using EcoRI and SpeI. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the restricted products and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. No colonies | ||
| + | |||
:https://static.igem.org/mediawiki/2012/0/00/W7_1_ara_restriktion.png | :https://static.igem.org/mediawiki/2012/0/00/W7_1_ara_restriktion.png | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 using EcoRI and SpeI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2 using EcoRI and SpeI | ||
| + | |||
* DH5a-cells containing the vectors A4.2_2, A3.1_2, A2.5_2, A5.13_5 were diluted in glycerol (20%) and freezed (-80°C). | * DH5a-cells containing the vectors A4.2_2, A3.1_2, A2.5_2, A5.13_5 were diluted in glycerol (20%) and freezed (-80°C). | ||
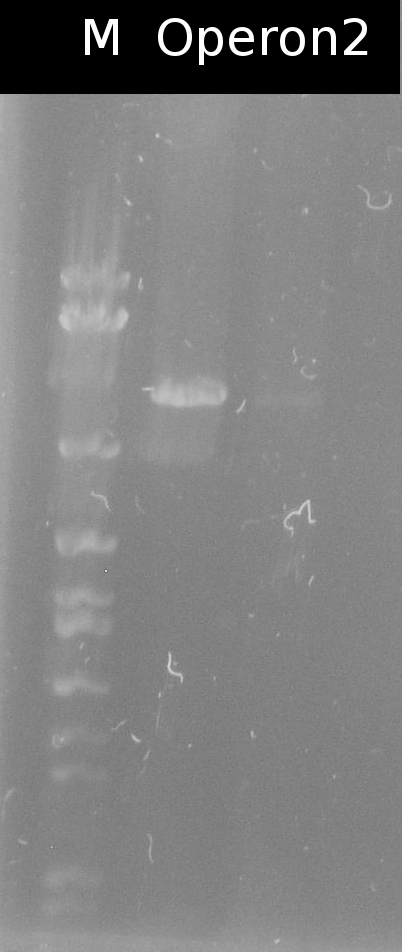
| - | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR Mutagenic PCR] of A4.2, A5.13, A3.1 to eliminate the PstI restriction site (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503#tctA503_Pst1 tctA_503_PstI], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162#tctB162_Pst1 tctB_162_PstI], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC#tphC_Pst1 tphC_PstI]) | + | |
| - | + | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR Mutagenic PCR] of A4.2, A5.13, A3.1 to eliminate the PstI restriction site (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503#tctA503_Pst1 tctA_503_PstI], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162#tctB162_Pst1 tctB_162_PstI], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC#tphC_Pst1 tphC_PstI]). [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] into DH5a | |
* Checking the DH5a-cells for correct transformation with the vectors after the mutagenic PCR by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (checking 7 colonies each, Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). The PCR products were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest digested] with PstI. The gel was alternately loaded with digested and uncut PCR products. | * Checking the DH5a-cells for correct transformation with the vectors after the mutagenic PCR by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (checking 7 colonies each, Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA503 tctA_503 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctB162 tctB_162 Biobrick], [https://2012.igem.org/Team:TU_Darmstadt/Materials/tphC tphC Biobrick]). The PCR products were [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest digested] with PstI. The gel was alternately loaded with digested and uncut PCR products. | ||
| + | |||
:https://static.igem.org/mediawiki/2012/b/b7/W7_2_trafo_mit_mut.png | :https://static.igem.org/mediawiki/2012/b/b7/W7_2_trafo_mit_mut.png | ||
| + | |||
* The transformation of DH5a with pSB1C3_tctA_505aa was verified by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]). The positive colonies were used for Miniprep | * The transformation of DH5a with pSB1C3_tctA_505aa was verified by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] (Primers: [https://2012.igem.org/Team:TU_Darmstadt/Materials/tctA505 tctA_505 Biobrick]). The positive colonies were used for Miniprep | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement: | * Miniprep (Fermentas Kit) and Nanodrop measurement: | ||
| + | |||
:A5.13m_1 : 105.2 ng/µl 260/280= 1.84 | :A5.13m_1 : 105.2 ng/µl 260/280= 1.84 | ||
| + | |||
:A5.13m_2 : 223.0 ng/µl 260/280= 1.87 | :A5.13m_2 : 223.0 ng/µl 260/280= 1.87 | ||
| + | |||
:A5.13m_3 : 110.6 ng/µl 260/280= 1.87 | :A5.13m_3 : 110.6 ng/µl 260/280= 1.87 | ||
| + | |||
:A5.13m_4 : 99.6 ng/µl 260/280= 1.90 | :A5.13m_4 : 99.6 ng/µl 260/280= 1.90 | ||
| + | |||
:A5.13m_6 : 326.6 ng/µl 260/280= 1.88 | :A5.13m_6 : 326.6 ng/µl 260/280= 1.88 | ||
| + | |||
:A5.13m_7 : 145.0 ng/µl 260/280= 1.84 | :A5.13m_7 : 145.0 ng/µl 260/280= 1.84 | ||
| + | |||
:A3.1m_1 : 241.7 ng/µl 260/280= 1.87 | :A3.1m_1 : 241.7 ng/µl 260/280= 1.87 | ||
| + | |||
:A3.1m_2 : 188.4 ng/µl 260/280= 1.89 | :A3.1m_2 : 188.4 ng/µl 260/280= 1.89 | ||
| + | |||
:A3.1m_3 : 112.4 ng/µl 260/280= 1.89 | :A3.1m_3 : 112.4 ng/µl 260/280= 1.89 | ||
| + | |||
:A3.1m_4 : 173.1 ng/µl 260/280= 1.85 | :A3.1m_4 : 173.1 ng/µl 260/280= 1.85 | ||
| + | |||
:A3.1m_5 : 111.6 ng/µl 260/280= 1.85 | :A3.1m_5 : 111.6 ng/µl 260/280= 1.85 | ||
| + | |||
:A3.1m_6 : 138.1 ng/µl 260/280= 1.87 | :A3.1m_6 : 138.1 ng/µl 260/280= 1.87 | ||
| + | |||
:A3.1m_7 : 178.9 ng/µl 260/280= 1.86 | :A3.1m_7 : 178.9 ng/µl 260/280= 1.86 | ||
| + | |||
:A4.2m_1 : 57.0 ng/µl 260/280= 1.84 | :A4.2m_1 : 57.0 ng/µl 260/280= 1.84 | ||
| + | |||
:A4.2m_2 : 46.5 ng/µl 260/280= 1.77 | :A4.2m_2 : 46.5 ng/µl 260/280= 1.77 | ||
| + | |||
:A4.2m_3 : 55.7 ng/µl 260/280= 1.88 | :A4.2m_3 : 55.7 ng/µl 260/280= 1.88 | ||
| + | |||
:A4.2m_4 : 111.3 ng/µl 260/280= 1.82 | :A4.2m_4 : 111.3 ng/µl 260/280= 1.82 | ||
| + | |||
:A4.2m_5 : 236.9 ng/µl 260/280= 1.89 | :A4.2m_5 : 236.9 ng/µl 260/280= 1.89 | ||
| + | |||
:A4.2m_6 : 69.3 ng/µl 260/280= 1.89 | :A4.2m_6 : 69.3 ng/µl 260/280= 1.89 | ||
| + | |||
:A4.2m_7 : 72.3 ng/µl 260/280= 1.85 | :A4.2m_7 : 72.3 ng/µl 260/280= 1.85 | ||
| + | |||
:C1.2 : 132.9 ng/µl 260/280= 1.72 | :C1.2 : 132.9 ng/µl 260/280= 1.72 | ||
| + | |||
* new [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of DH5a with pXylE_Ara | * new [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of DH5a with pXylE_Ara | ||
| + | |||
| + | |||
===week 8 (09.-13.07.12)=== | ===week 8 (09.-13.07.12)=== | ||
| + | |||
* Checking the transformation of DH5a with pSB1C3_tctA_505aa again ([https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]) [*]. The colonies Nr. 2, 8 and 10 were picked and streaked onto fresh media. | * Checking the transformation of DH5a with pSB1C3_tctA_505aa again ([https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]) [*]. The colonies Nr. 2, 8 and 10 were picked and streaked onto fresh media. | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of DH5a_pXylE_Ara [*] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of DH5a_pXylE_Ara [*] | ||
| + | |||
:No positive colonies | :No positive colonies | ||
| + | |||
* Sequencing the following vectors: pSB1A2_4.2m_5, pSB1A2_4.2m_4, pSB1A2_5.13m_3, pSB1A2_5.13m_2, pSB1A2_3.1m_1, pSB1A2_3.1m_5, pSB1A2_3.1m_7 | * Sequencing the following vectors: pSB1A2_4.2m_5, pSB1A2_4.2m_4, pSB1A2_5.13m_3, pSB1A2_5.13m_2, pSB1A2_3.1m_1, pSB1A2_3.1m_5, pSB1A2_3.1m_7 | ||
| + | |||
* Checking the transformation of DH5a with A1 (5 colonies) and C1 (7 colonies) by using Colony-PCR [?] | * Checking the transformation of DH5a with A1 (5 colonies) and C1 (7 colonies) by using Colony-PCR [?] | ||
| + | |||
* Checking the transformation of DH5a with pSB1C3_tctA_505aa again by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]. Only the colonies Nr. 2, 8 and 10 (see above) were checked (2 colonies each). | * Checking the transformation of DH5a with pSB1C3_tctA_505aa again by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]. Only the colonies Nr. 2, 8 and 10 (see above) were checked (2 colonies each). | ||
| + | |||
:https://static.igem.org/mediawiki/2012/f/f0/W8_3_kolo_pcr_505.png | :https://static.igem.org/mediawiki/2012/f/f0/W8_3_kolo_pcr_505.png | ||
| + | |||
* Another [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of DH5a_pXy19_Ara | * Another [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] of DH5a_pXy19_Ara | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:A5.13m_2 : 191.2 ng/µl 260/280= 1.84 | :A5.13m_2 : 191.2 ng/µl 260/280= 1.84 | ||
| + | |||
:A5.13m_3 : 168.1 ng/µl 260/280= 1.78 | :A5.13m_3 : 168.1 ng/µl 260/280= 1.78 | ||
| + | |||
:A2.5 : 193.0 ng/µl 260/280= 1.83 | :A2.5 : 193.0 ng/µl 260/280= 1.83 | ||
| + | |||
:A3.1m_5 : 146.1 ng/µl 260/280= 1.82 | :A3.1m_5 : 146.1 ng/µl 260/280= 1.82 | ||
| + | |||
:A3.1m_1 : 250.1 ng/µl 260/280= 1.84 | :A3.1m_1 : 250.1 ng/µl 260/280= 1.84 | ||
| + | |||
:A4.2m_4 : 124.8 ng/µl 260/280= 1.85 | :A4.2m_4 : 124.8 ng/µl 260/280= 1.85 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of A5.13m_2, A5.13m_3, A3.1m_1, A3.1m_5, A2.5, A4.2m_4 with EcoRI and SpeI. The restriction was done to enable the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] of the mutated genes into pSB1C3. Afterwards DH5a was [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformed] with the ligation product. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of A5.13m_2, A5.13m_3, A3.1m_1, A3.1m_5, A2.5, A4.2m_4 with EcoRI and SpeI. The restriction was done to enable the [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] of the mutated genes into pSB1C3. Afterwards DH5a was [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformed] with the ligation product. | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:C505_2_2 : 253.6 ng/µl 260/280= 1.60 | :C505_2_2 : 253.6 ng/µl 260/280= 1.60 | ||
| + | |||
:C505_2_3 : 274.7 ng/µl 260/280= 1.88 | :C505_2_3 : 274.7 ng/µl 260/280= 1.88 | ||
| + | |||
:C505_8_2 : 462.1 ng/µl 260/280= 1.38 | :C505_8_2 : 462.1 ng/µl 260/280= 1.38 | ||
| + | |||
:C505_8_3 : 149.1 ng/µl 260/280= 1.84 | :C505_8_3 : 149.1 ng/µl 260/280= 1.84 | ||
| + | |||
:C505_10_3 : 160.4 ng/µl 260/280= 1.98 | :C505_10_3 : 160.4 ng/µl 260/280= 1.98 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the Miniprep products with EcoRI and EcoRI/SpeI. Afterwards the restriction products were analyzed by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Gel loading: uncut, cut once, cut twice | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the Miniprep products with EcoRI and EcoRI/SpeI. Afterwards the restriction products were analyzed by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis]. Gel loading: uncut, cut once, cut twice | ||
| + | |||
:https://static.igem.org/mediawiki/2012/7/7e/W8_4_restriktion_505.png | :https://static.igem.org/mediawiki/2012/7/7e/W8_4_restriktion_505.png | ||
| + | |||
:C505_2_3 und C505_8_2 were selected for mutagenic PCR. | :C505_2_3 und C505_8_2 were selected for mutagenic PCR. | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3 with EcoRI and SpeI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3 with EcoRI and SpeI | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pXylE with EcoRI and XbaI and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] with EcoRI and SpeI. Dephosphorylation of pXylE. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of pXylE and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pXylE with EcoRI and XbaI and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] with EcoRI and SpeI. Dephosphorylation of pXylE. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of pXylE and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] | ||
| + | |||
| + | |||
===week 9 (16.-20.07.12)=== | ===week 9 (16.-20.07.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR mutagenic PCR] of C505_2_3 and C505_8_2. (Did not work) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR mutagenic PCR] of C505_2_3 and C505_8_2. (Did not work) | ||
| + | |||
* [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tct_B162m] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tct_B197m] with primer to add a RBS in front and behind the gene | * [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tct_B162m] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tct_B197m] with primer to add a RBS in front and behind the gene | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the PCR products with EcoRI and SpeI for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] into pSB1C3_5.13m (tphC) | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of the PCR products with EcoRI and SpeI for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] into pSB1C3_5.13m (tphC) | ||
| + | |||
* Adding a RBS to one site of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] using PCR with the reverse primer | * Adding a RBS to one site of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] using PCR with the reverse primer | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:C3.1m_1 : 278.9 ng/µl 260/280= 1.88 | :C3.1m_1 : 278.9 ng/µl 260/280= 1.88 | ||
| + | |||
:C4.2m_4 : 214.2 ng/µl 260/280= 1.86 | :C4.2m_4 : 214.2 ng/µl 260/280= 1.86 | ||
| + | |||
:5.13m_3 : 256.3 ng/µl 260/280= 1.88 | :5.13m_3 : 256.3 ng/µl 260/280= 1.88 | ||
| + | |||
:C2.5_2 : 44.8 ng/µl 260/280= 2.00 | :C2.5_2 : 44.8 ng/µl 260/280= 2.00 | ||
| + | |||
:C5.13m_2 : 60.8 ng/µl 260/280= 1.89 | :C5.13m_2 : 60.8 ng/µl 260/280= 1.89 | ||
| + | |||
| + | |||
===week 10 (23.-27.07.12)=== | ===week 10 (23.-27.07.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_5.13m_3_2 and pSB1C3_5.13m_3_3 with EcoRI and XbaI. Afterwards the products are [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligated] with RBS_tctB162m_RBS and RBS_tctB197_RBS each. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_5.13m_3_2 and pSB1C3_5.13m_3_3 with EcoRI and XbaI. Afterwards the products are [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligated] with RBS_tctB162m_RBS and RBS_tctB197_RBS each. | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of A3.1m_1 for [[ligation]] with Ara | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of A3.1m_1 for [[ligation]] with Ara | ||
| + | |||
* [[Digestion]] of pXylE for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] with Ara_RBS | * [[Digestion]] of pXylE for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] with Ara_RBS | ||
| + | |||
* [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of Ara_RBS and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] into pXylE and pSB1C3. Afterwards [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest restriction digest] with restriction enzymes: Ara_RBS is cut with with EcoRI and SpeI and Ara is cut with EcoRI and XbaI | * [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of Ara_RBS and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] for [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] into pXylE and pSB1C3. Afterwards [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest restriction digest] with restriction enzymes: Ara_RBS is cut with with EcoRI and SpeI and Ara is cut with EcoRI and XbaI | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the restricted PCR-products to: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of the restricted PCR-products to: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR mutagenic PCR] of 505_8_2 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR mutagenic PCR] of 505_8_2 | ||
| + | |||
| + | |||
===week 11 (30.07.-03.08.12)=== | ===week 11 (30.07.-03.08.12)=== | ||
| + | |||
* Checking [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of DH5a by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE. Positive colonies are picked and used for inoculation of fluid media. | * Checking [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of DH5a by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR]: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE. Positive colonies are picked and used for inoculation of fluid media. | ||
| + | |||
* Miniprep (Fermentas Kit) of fluid culture and Nanodrop measurement of: | * Miniprep (Fermentas Kit) of fluid culture and Nanodrop measurement of: | ||
| + | |||
:pSB1C3_503 : 110.0 ng/µl 260/280= 1.82 | :pSB1C3_503 : 110.0 ng/µl 260/280= 1.82 | ||
| + | |||
:pSB1A2_tctB197_tphCm : 32.0 ng/µl 260/280= 2.10 | :pSB1A2_tctB197_tphCm : 32.0 ng/µl 260/280= 2.10 | ||
| + | |||
:pSB1A2_Ara_tctA503m : 202.0 ng/µl 260/280= 1.78 | :pSB1A2_Ara_tctA503m : 202.0 ng/µl 260/280= 1.78 | ||
| + | |||
:pSB1A2_tctB162m_tphCm : 36.5 ng/µl 260/280= 1.91 | :pSB1A2_tctB162m_tphCm : 36.5 ng/µl 260/280= 1.91 | ||
| + | |||
:pSB1C3_Ara_tctA503m : 133.2 ng/µl 260/280= 1.83 | :pSB1C3_Ara_tctA503m : 133.2 ng/µl 260/280= 1.83 | ||
| + | |||
:pSB1C3_tctB162m_tphCm : 35.2 ng/µl 260/280= 1.93 | :pSB1C3_tctB162m_tphCm : 35.2 ng/µl 260/280= 1.93 | ||
| + | |||
:pSB1C3_tct197_tphC : 30.6 ng/µl 260/280= 1.88 | :pSB1C3_tct197_tphC : 30.6 ng/µl 260/280= 1.88 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm and building the final constructs by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation]: pSB1A2_tctA503m_tctB162m_tphCm und pSB1C3_tctA503m_tctB162m_tphCm. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of the final constructs into DH5a. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm and building the final constructs by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation]: pSB1A2_tctA503m_tctB162m_tphCm und pSB1C3_tctA503m_tctB162m_tphCm. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of the final constructs into DH5a. | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of pSB1C3_503 into DH5a | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation Transformation] of pSB1C3_503 into DH5a | ||
| + | |||
* Checking the mutagenic PCR product of 505_8_2 and the transformation of DH5a with pSB1C3_503 by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] | * Checking the mutagenic PCR product of 505_8_2 and the transformation of DH5a with pSB1C3_503 by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] und C4.2_4_1 using EcoRI and PstI, building pSB1C3_Ara by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of the vector into DH5a | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] und C4.2_4_1 using EcoRI and PstI, building pSB1C3_Ara by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] of the vector into DH5a | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:pSB1C3_Ara : 86.7 ng/µl 260/280= 1.83 | :pSB1C3_Ara : 86.7 ng/µl 260/280= 1.83 | ||
| + | |||
:pSB1C3_505m : 113.8 ng/µl 260/280= 1.85 | :pSB1C3_505m : 113.8 ng/µl 260/280= 1.85 | ||
| + | |||
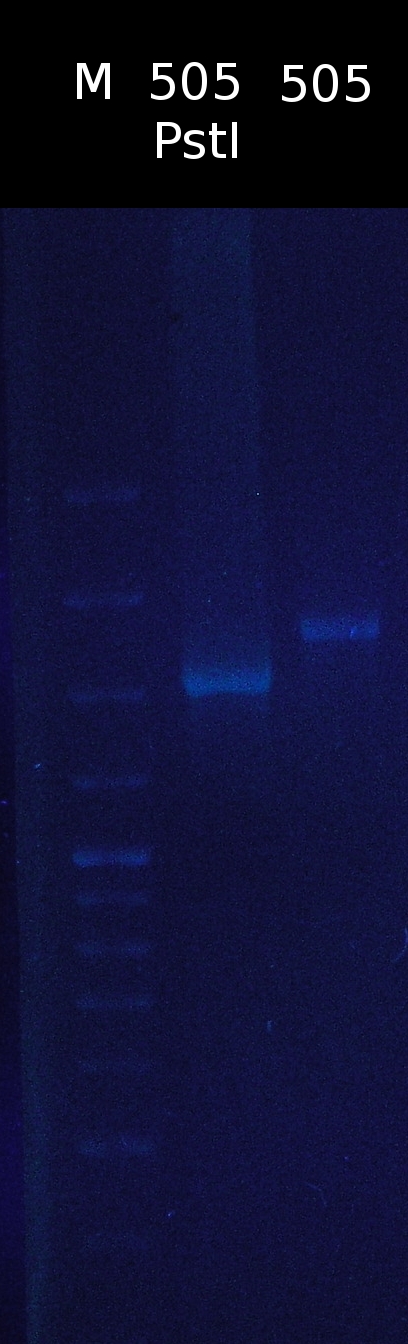
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_505m with PstI. Checking if the restriction site has been successfully removed | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_505m with PstI. Checking if the restriction site has been successfully removed | ||
| + | |||
:[[File:W11_1_rest_mut_505.jpg|100px]] | :[[File:W11_1_rest_mut_505.jpg|100px]] | ||
| + | |||
:restriction site hasn't been removed | :restriction site hasn't been removed | ||
| + | |||
* Screnning for transformation of DH5a with pSB1A2_tctA503m_tctB162m_tphCm and pSB1C3_tctA503m_tctB162m_tphCm using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] [*] | * Screnning for transformation of DH5a with pSB1A2_tctA503m_tctB162m_tphCm and pSB1C3_tctA503m_tctB162m_tphCm using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] [*] | ||
| + | |||
* Checking the constructs pSB1A2/pSB1C3_Ara_503m and pSB1A2/pSB1C3_tctB162m_tphCm by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] [*] | * Checking the constructs pSB1A2/pSB1C3_Ara_503m and pSB1A2/pSB1C3_tctB162m_tphCm by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] [*] | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] into pSB1C3 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] into pSB1C3 | ||
| + | |||
* Checking if pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm have the right Prefix/Suffix by digesting both with EcoRI and XbaI and with EcoRI and PstI [*] | * Checking if pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm have the right Prefix/Suffix by digesting both with EcoRI and XbaI and with EcoRI and PstI [*] | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm with Ara_503 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm with Ara_503 | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:pSB1A2_tctB162m_tphCm_1 : 45.3 ng/µl 260/280= 1.90 | :pSB1A2_tctB162m_tphCm_1 : 45.3 ng/µl 260/280= 1.90 | ||
| + | |||
:pSB1A2_tctB162m_tphCm_2 : 74.5 ng/µl 260/280= 1.82 | :pSB1A2_tctB162m_tphCm_2 : 74.5 ng/µl 260/280= 1.82 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_1 : 95.8 ng/µl 260/280= 1.86 | :pSB1C3_tctB162m_tphCm_1 : 95.8 ng/µl 260/280= 1.86 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_2 : 85.7 ng/µl 260/280= 1.81 | :pSB1C3_tctB162m_tphCm_2 : 85.7 ng/µl 260/280= 1.81 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_3 : 100.1 ng/µl 260/280= 1.84 | :pSB1C3_tctB162m_tphCm_3 : 100.1 ng/µl 260/280= 1.84 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_4 : 79.9 ng/µl 260/280= 1.88 | :pSB1C3_tctB162m_tphCm_4 : 79.9 ng/µl 260/280= 1.88 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_5 : 89.8 ng/µl 260/280= 1.85 | :pSB1C3_tctB162m_tphCm_5 : 89.8 ng/µl 260/280= 1.85 | ||
| + | |||
* Sequencing pSB1A2_tctB162m_tphCm_2 and pSB1C3_tctB162m_tphCm_5 | * Sequencing pSB1A2_tctB162m_tphCm_2 and pSB1C3_tctB162m_tphCm_5 | ||
| + | |||
| + | |||
===week 12 (06.-10.08.12)=== | ===week 12 (06.-10.08.12)=== | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:pSB1A2_tctB162m_tphCm_2 : 55.7 ng/µl 260/280= 1.81 | :pSB1A2_tctB162m_tphCm_2 : 55.7 ng/µl 260/280= 1.81 | ||
| + | |||
:pSB1C3_tctB162m_tphCm_5 : 101.2 ng/µl 260/280= 1.92 | :pSB1C3_tctB162m_tphCm_5 : 101.2 ng/µl 260/280= 1.92 | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR Mutagenic PCR] of pSB1C3_505_8_2 and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/mutagenic_PCR Mutagenic PCR] of pSB1C3_505_8_2 and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of Ara_503m with pSB1A2_tctB162m_tphCm/pSB1C3_tctB162m_tphCm and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. Screening the transformants using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of Ara_503m with pSB1A2_tctB162m_tphCm/pSB1C3_tctB162m_tphCm and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. Screening the transformants using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | ||
| + | |||
:[[File:W12_1_kolo_operon1.JPG|250px]] | :[[File:W12_1_kolo_operon1.JPG|250px]] | ||
| + | |||
:only colony 1, 2 and 3 have the right insert | :only colony 1, 2 and 3 have the right insert | ||
| + | |||
* Miniprep (Fermentas Kit) and Nanodrop measurement of: | * Miniprep (Fermentas Kit) and Nanodrop measurement of: | ||
| + | |||
:pSB1A2_Ara_tctA503m : 278.0 ng/µl 260/280= 1.86 | :pSB1A2_Ara_tctA503m : 278.0 ng/µl 260/280= 1.86 | ||
| + | |||
:pSB1C3_Ara_tctA503m : 169.0 ng/µl 260/280= 1.87 | :pSB1C3_Ara_tctA503m : 169.0 ng/µl 260/280= 1.87 | ||
| + | |||
:pSB1C3_Ara_1 : 59.0 ng/µl 260/280= 1.90 | :pSB1C3_Ara_1 : 59.0 ng/µl 260/280= 1.90 | ||
| + | |||
:pSB1C3_Ara_4 : 55.5 ng/µl 260/280= 1.80 | :pSB1C3_Ara_4 : 55.5 ng/µl 260/280= 1.80 | ||
| + | |||
:pSB1C3_Ara_tctA503m_tctB162m_tphCm : 188.8 ng/µl 260/280= 1.86 | :pSB1C3_Ara_tctA503m_tctB162m_tphCm : 188.8 ng/µl 260/280= 1.86 | ||
| + | |||
:pSB1C3_Ara_tctA503m_tctB162m_tphCm :168.8 ng/µl 260/280= 1.84 | :pSB1C3_Ara_tctA503m_tctB162m_tphCm :168.8 ng/µl 260/280= 1.84 | ||
| + | |||
:pSB1C3_Ara_tctA503m_tctB162m_tphCm :271.3 ng/µl 260/280= 1.85 | :pSB1C3_Ara_tctA503m_tctB162m_tphCm :271.3 ng/µl 260/280= 1.85 | ||
| + | |||
:pSB1C3_Ara_XylE : 149.8 ng/µl 260/280= 1.86 | :pSB1C3_Ara_XylE : 149.8 ng/µl 260/280= 1.86 | ||
| + | |||
:pSB1C3_tctB197_tphC : 95.8 ng/µl 260/280= 1.84 | :pSB1C3_tctB197_tphC : 95.8 ng/µl 260/280= 1.84 | ||
| + | |||
:pSB1A2_tctB197_tphC : 69.0 ng/µl 260/280= 1.88 | :pSB1A2_tctB197_tphC : 69.0 ng/µl 260/280= 1.88 | ||
| + | |||
* Sequencing of 3 different pSB1C3_Ara_tctA503m_tctB162m_tphC | * Sequencing of 3 different pSB1C3_Ara_tctA503m_tctB162m_tphC | ||
| + | |||
* Analysis of mutagenic PCR products of 505_8_2 and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest restriction digest] with PstI | * Analysis of mutagenic PCR products of 505_8_2 and [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest restriction digest] with PstI | ||
| + | |||
:[[File:W12_2_505m_restriktion.JPG|400px]] | :[[File:W12_2_505m_restriktion.JPG|400px]] | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of Ara_RBS with EcoRI and SpeI and of pSB1C3_505m with EcoRI and XbaI. Building pSB1C3_Ara_505m by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of Ara_RBS with EcoRI and SpeI and of pSB1C3_505m with EcoRI and XbaI. Building pSB1C3_Ara_505m by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a. | ||
| + | |||
| + | |||
===week 13 (13.-17.08.12)=== | ===week 13 (13.-17.08.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_197_tphCm with EcoRI and XbaI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_197_tphCm with EcoRI and XbaI | ||
| + | |||
* Checking the transformation of DH5a with pSB1C3_Ara_505m by using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | * Checking the transformation of DH5a with pSB1C3_Ara_505m by using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | ||
| + | |||
:[[File:W13_1_505_mut_PCR_von_Basti.JPG|500px]] | :[[File:W13_1_505_mut_PCR_von_Basti.JPG|500px]] | ||
| + | |||
* Sequencing pSB1C3_Ara_XylE, pSB1C3_Ara_tctA503m_tctB162m_tphCm (=Operon1), pSB1C3_505m | * Sequencing pSB1C3_Ara_XylE, pSB1C3_Ara_tctA503m_tctB162m_tphCm (=Operon1), pSB1C3_505m | ||
| + | |||
*[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of Ara_RBS with EcoRI and SpeI and of pSB1C3_tphCm with EcoRI and XbaI. Building pSB1C3_Ara_tphCm by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | *[https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of Ara_RBS with EcoRI and SpeI and of pSB1C3_tphCm with EcoRI and XbaI. Building pSB1C3_Ara_tphCm by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation] and subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_Ara_tctA505m with EcoRI and SpeI. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of Ara_tctA505m and tctB197_tphCm into pSB1C3 | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_Ara_tctA505m with EcoRI and SpeI. [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation Ligation] of Ara_tctA505m and tctB197_tphCm into pSB1C3 | ||
| + | |||
| + | |||
===week 14 (20.-24.08.12)=== | ===week 14 (20.-24.08.12)=== | ||
| + | |||
* Checking the transformation of DH5a with pSB1C3_Ara_tphCm using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | * Checking the transformation of DH5a with pSB1C3_Ara_tphCm using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | ||
| + | |||
:[[File:W14_1_kolo.JPG|250px]] | :[[File:W14_1_kolo.JPG|250px]] | ||
| + | |||
| + | |||
===week 15 (27.-31.08.12)=== | ===week 15 (27.-31.08.12)=== | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_Ara_tphCm and pSB1C3_Ara_tctA505m with PstI and SpeI | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_Ara_tphCm and pSB1C3_Ara_tctA505m with PstI and SpeI | ||
| + | |||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_197_tphCm with XbaI and PstI. | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Restriction_digest Restriction digest] of pSB1C3_197_tphCm with XbaI and PstI. | ||
| + | |||
* Building the construct pSB1C3_Ara_tctA505m_197_tphCm (= Operon2) by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation]. Subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | * Building the construct pSB1C3_Ara_tctA505m_197_tphCm (= Operon2) by [https://2012.igem.org/Team:TU_Darmstadt/Protocols/DNA_Ligation ligation]. Subsequent [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Bacterial_transformation transformation] into DH5a | ||
| + | |||
* Checking the transformation of DH5a with Operon2 using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | * Checking the transformation of DH5a with Operon2 using [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Colony_PCR colony-PCR] | ||
| + | |||
:[[File:W15_1_operon2_Kolo.jpg|100px]] | :[[File:W15_1_operon2_Kolo.jpg|100px]] | ||
Revision as of 16:48, 18 September 2012
Transport
Week 1 (21.-25.05.12)
- First colony PCR of Comamonas testosteroni for isolation of the following genes: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5). (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick). Analysis of [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] products by Agarose gel electrophoresis
- PCR product purification using the Promega-Kit
- Nanodrop measurements of the purified PCR products:
- (1) : 19.5 ng/µl 260/280=1.58
- (2) : 61.9 ng/µl 260/280=1.75
- (3) : 75.1 ng/µl 260/280=1.75
- (4) : 93.3 ng/µl 260/280=1.85
- (5) : 91.3 ng/µl 260/280=1.58
- Restriction digest of PCR products by SpeI and EcoRI, heat inactivation after digestion
- Analysis of restriction digest by Agarose gel electrophoresis
- Purification of the bands gained form gel electrophoresis (Promega-Kit)
- Nanodrop measurements of the purified products:
- (1) : 8.3 ng/µl 260/280=1.94
- (2) : 8.9 ng/µl 260/280=1.80
- (3) : 13.0 ng/µl 260/280=1.57
- (4) : 1.5 ng/µl 260/280=3.08
- (5) : 3.7 ng/µl 260/280=2.26
week 2 (28.05.-01.06.12)
- Purified products from 1. week were ligated into pSB1A2 and subsequently transformed into DH5a (in spite of low concentration)
- Overnight incubation on LB agar plates; no growth detectable
- Second approach to isolate the five genes from Comamonas t. using colony-PCR (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick)
- Analysis of PCR products by Agarose gel electrophoresis; without results; modification of the PCR protocols: adding DMSO
- Third approach to isolate the five genes from Comamonas t. by colony-PCR (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick)
- PCR product purification using the Promega-Kit: ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5))
- (1): no PCR-product
- Nanodrop measurements of the purified PCR products:
- (2) : 56.1 ng/µl 260/280=1.87
- (3) : 66.8 ng/µl 260/280=1.92
- (4) : 72.8 ng/µl 260/280=1.87
- (5) : 68.8 ng/µl 260/280=1.82
week 3 (04.-08.06.12)
- Restriction digest of the purified PCR products and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (leftover of the 1. week) by SpeI and EcoRI
- Restriction digest of pSB1A2 using SpeI and EcoRI
- Dephosphorylation of the restriction reactions by using antarctic Phosphatase
- Ligation of the five genes into pSB1A2 and subsequently transformation into DH5a (incubation at 37°C)
- colony-PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches (Primer:tctA_505 Biobrick)
- Analysis of PCR products by Agarose gel electrophoresis. Only one of them showed the expected band. The band was excluded and purified via the Promega-Kit.
- Nanodrop measurement of the purified product:
- (1.1) =8.9 ng/µl 260/280=2,22
- Because of low concentration and contamination the PCR was repeated and the product was purified again by the Promega-Kit.
- Performing a colony-PCR-screen of the transformed cells using the Taq-Polymerase. Verification of 4 colonies per plate. (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick)
- PCR did not work
- Testing the Taq-Polymerase for function. The Phu-Polymerase was used for comparison. Only the approach using the Phu-Polymerase showed the expected bands in Agarose gel electrophoresis.
- Screening was repeated with Phu-Polymerase; no bands visible
week 4 (11.-15-06.12)
- Inoculation of LB-media with DH5a_pSB1A2 and DH5a_pSB1C3; overnight incubation at 37°C
- Miniprep of DH5a_pSB1A2 and DH5a_pSB1C3 using the Promega-Kit
- Nanodrop measurements of the preparation:
- pSB1A2 : 136.1 ng/µl 260/280= 1.93
- pSB1C3 : 265.1 ng/µl 260/280= 1.89
- Restriction digest of pSB1A2 and pSB1C3 with EcoRI and SpeI. Afterwards dephosphorylation by using Antarctic phosphatase.
- Analysis of the restriction products through Agarose gel electrophoresis; for comparison the uncropped vectors were also analyzed.
- pSB1C3 shows several bands (Insert, vector without insert, linearized vector with insert); concentration of pSB1A2 is very low, only one band visible
- Approach to isolate the following genes from Comamonas t. by colony-PCR: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick)
- Analysis of PCR products by Agarose gel electrophoresis
- Isolation of (1) and (5) did not work
- PCR product purification by using the Promega-Kit
- Nanodrop measurements of the purified PCR products:
- (2) : 18.3 ng/µl 260/280=2.03
- (3) : 21.2 ng/µl 260/280=1.95
- (4) : 23.9 ng/µl 260/280=2.05
- Restriction digest of purified PCR products (2), (3) and (4) using SpeI and EcoRI
- Ligation of the digested genes ((2), (3), (4)) into pSB1A2 and pSB1C3
- Transformation of DH5a with pSB1A2_197, pSB1A2_503, pSB1A2_162, pSB1C3_197, pSB1C3_503, pSB1C3_162 (6 different transformation approaches). Overnight incubation at 37°C
- Screening of the transformants by colony-PCR (Primer: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick). Verification of 6 colonies for every transformation. Afterwards the PCR products were analyzed by Agarose gel electrophoresis
- positive colonies are marked
- Colony-PCR of Comamonas t. to isolate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) und [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) (Primer: tctA_505 Biobrick, tphC Biobrick)
- Analysis of PCR products (Colony-PCR) and the ligation approach by Agarose gel electrophoresis
- PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] shows only the band of the undesired smaller product
- Restriction digest of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) and subsequent Ligation into pSB1A2 and pSB1C3
- Transformation of DH5a with pSB1A2_322 und pSB1C3_322. [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primer: tphC Biobrick) used to check for successful transformation
- Isolation of Comamonas t. genome, because it seems impossible to amplificate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) from a fluid Comamonas t. culture.
week 5 (18.-22.06.12)
- Introduction of a nomenclature: A/C gene/colony: pSB1A2 = A; pSB1C3 = C; gene: tctA 505aa = 1, tctB 197aa = 2, tctA 503aa = 3, tctB 162aa = 4, tphC 322aa = 5; colony = 1...X
- Producing fluid cultures of the positive transformants
- Miniprep of the fluid culures by using the Frementas-Kit. Afterwards Nanodrop measurements of the preparations
- A2.3 : 125.9 ng/µl 260/280= 1.62
- A3.1 : 103.3 ng/µl 260/280= 1.72
- A4.6 : 147.2 ng/µl 260/280= 1.70
- A4.5 : 108.7 ng/µl 260/280= 1.86
- A5.12 : 88.0 ng/µl 260/280= 1.87
- A5.8 : 85.01 ng/µl 260/280= 1.78
- A3.2 : 87.3 ng/µl 260/280= 1.81
- C4.6 : 70.5 ng/µl 260/280= 1.81
- C4.1 : 41.2 ng/µl 260/280= 1.85
- C4.4 : 104.2 ng/µl 260/280= 1.85
- C2.1 : 132.2 ng/µl 260/280= 1.85
- C2.6 : 188.1 ng/µl 260/280= 1.75
- [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] using a rest of purified [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1) (Primer: tctA_505 Biobrick), subsequent Agarose gel electrophoresis
- Another colony-PCR using the transformants (2)-(5) as template (Primers: tctA_503 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick); analyzing the PCR product by Agarose gel electrophoresis
- 3 positive transformants were picked for inoculation of fluid media (A34, A43; C513)
- Miniprep (Fermentas Kit) of the fluid culures and Nanodrop measurements:
- A2.4 : 202,1 ng/µl 260/280= 1,63
- A2.5 : 126,5 ng/µl 260/280= 1,90
- A2.6 : 107,5 ng/µl 260/280= 1,81
- A4.2 : 133,8 ng/µl 260/280= 1,59
- C2.3 : 187,8 ng/µl 260/280= 1,87
- C2.4 : 270,7 ng/µl 260/280= 1,80
- C4.1 : 103,8 ng/µl 260/280= 1,80
- The Miniprep products were Restriction digested by restriction enzymes, each was cut once with EcoRI and twice with EcoRI/SpeI. Analisys of the digested products by [Agarose gel electrophoresis
week 6 (25.-29.06.12)
- Further colony-PCR of Comamonas t. (3 approaches) and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic Comamonas t. DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1), 2 approaches each (Primer: tctA_505 Biobrick). Subsequent analisys of the PCR products by Agarose gel electrophoresis. All PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] show only the band of the undesired smaller product.
- Ligation off the digested genes into pSB1A2 and transformation into DH5a. Overnight incubation at 37°C
- Colony-PCR of Comamonas t. and [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] with genomic Comamonas t. DNA as template to obtain [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] (1). [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tctB 197aa] (2), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808002 tctA_503aa] (3), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tctB 162aa] (4), [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808001 tphC 322aa] (5) used as control (Primers: tctA_503 Biobrick, tctA_505 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick)
- Miniprep and Nanodrop measurement of:
- A3.1 : 111.6 ng/µl 260/280= 1.72
- A3.4 : 79.5 ng/µl 260/280= 1.66
- C4.6 : 69.6 ng/µl 260/280= 1.76
- A4.2 : 96.9 ng/µl 260/280= 1.62
- A2.4 : 109.7 ng/µl 260/280= 1.72
- A2.5 : 160.8 ng/µl 260/280= 1.79
- A5.13 : 188.7 ng/µl 260/280= 1.85
- A5.12 : 114.5 ng/µl 260/280= 1.81
- Checking the Miniprep with [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] (Primers: tctA_503 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick) and Agarose gel electrophoresis
- Colony-PCR-Screen of the transformants (Primer: tctA_505 Biobrick) and analisys of the PCR products by Agarose gel electrophoresis
- No bands visible
- Ligation of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] into pSB1C3 and subsequently transformation into DH5a. No colonies
- The following vectors were sequenced: pSB1A2_197.4, pSB1A2_197.5, psSB1A2_503.1, pSB1A2_503.4, pSB1A2_162.2, pSB1C3_162.6, pSB1A2_322.12, pSB1A2_322.13
- Restriction digest of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808004 tctA 505aa] using EcoRI/SpeI. Ligation into pSB1C3 and subsequently transformation into DH5a.
- Preperations for pure culture: colony-PCR of A2.4, A2.5, A3.1, A3.4, A4.2, C4.6, A5.12, A5.13 (2 colonies each, Primers: tctA_503 Biobrick, tctB_162 Biobrick, tctB_197 Biobrick, tphC Biobrick) and afterwards checking the PCR products by Agarose gel electrophoresis. Positive colonies were picked, streaked onto fresh media and incubated again.
- As A5.12 and A5.13 are missing the right insert the colony-PCR was repeated (5 and 6 colonies were used, Primer: tphC Biobrick). To provide additional monitoring the colony-PCR was repeated for A2.5_2, A3.1_2 und A4.2_2. 2 positive colonies were again streaked onto fresh media.
- The Arabinose-Promotor ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara]) was isolated from pbad_turbo_gfp and colonies containing this plasmid (3 approaches each). Approach T2 is further in use.
week 7 (02.-06.07.12)
- Restriction digest of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] and pXylE using EcoRI and SpeI. Ligation of the restricted products and transformation into DH5a. No colonies
- Restriction digest of pSB1A2 using EcoRI and SpeI
- DH5a-cells containing the vectors A4.2_2, A3.1_2, A2.5_2, A5.13_5 were diluted in glycerol (20%) and freezed (-80°C).
- Mutagenic PCR of A4.2, A5.13, A3.1 to eliminate the PstI restriction site (Primers: tctA_503_PstI, tctB_162_PstI, tphC_PstI). Transformation into DH5a
- Checking the DH5a-cells for correct transformation with the vectors after the mutagenic PCR by colony-PCR (checking 7 colonies each, Primers: tctA_503 Biobrick, tctB_162 Biobrick, tphC Biobrick). The PCR products were digested with PstI. The gel was alternately loaded with digested and uncut PCR products.
- The transformation of DH5a with pSB1C3_tctA_505aa was verified by colony-PCR (Primers: tctA_505 Biobrick). The positive colonies were used for Miniprep
- Miniprep (Fermentas Kit) and Nanodrop measurement:
- A5.13m_1 : 105.2 ng/µl 260/280= 1.84
- A5.13m_2 : 223.0 ng/µl 260/280= 1.87
- A5.13m_3 : 110.6 ng/µl 260/280= 1.87
- A5.13m_4 : 99.6 ng/µl 260/280= 1.90
- A5.13m_6 : 326.6 ng/µl 260/280= 1.88
- A5.13m_7 : 145.0 ng/µl 260/280= 1.84
- A3.1m_1 : 241.7 ng/µl 260/280= 1.87
- A3.1m_2 : 188.4 ng/µl 260/280= 1.89
- A3.1m_3 : 112.4 ng/µl 260/280= 1.89
- A3.1m_4 : 173.1 ng/µl 260/280= 1.85
- A3.1m_5 : 111.6 ng/µl 260/280= 1.85
- A3.1m_6 : 138.1 ng/µl 260/280= 1.87
- A3.1m_7 : 178.9 ng/µl 260/280= 1.86
- A4.2m_1 : 57.0 ng/µl 260/280= 1.84
- A4.2m_2 : 46.5 ng/µl 260/280= 1.77
- A4.2m_3 : 55.7 ng/µl 260/280= 1.88
- A4.2m_4 : 111.3 ng/µl 260/280= 1.82
- A4.2m_5 : 236.9 ng/µl 260/280= 1.89
- A4.2m_6 : 69.3 ng/µl 260/280= 1.89
- A4.2m_7 : 72.3 ng/µl 260/280= 1.85
- C1.2 : 132.9 ng/µl 260/280= 1.72
- new transformation of DH5a with pXylE_Ara
week 8 (09.-13.07.12)
- Checking the transformation of DH5a with pSB1C3_tctA_505aa again (colony-PCR) [*]. The colonies Nr. 2, 8 and 10 were picked and streaked onto fresh media.
- colony-PCR of DH5a_pXylE_Ara [*]
- No positive colonies
- Sequencing the following vectors: pSB1A2_4.2m_5, pSB1A2_4.2m_4, pSB1A2_5.13m_3, pSB1A2_5.13m_2, pSB1A2_3.1m_1, pSB1A2_3.1m_5, pSB1A2_3.1m_7
- Checking the transformation of DH5a with A1 (5 colonies) and C1 (7 colonies) by using Colony-PCR [?]
- Checking the transformation of DH5a with pSB1C3_tctA_505aa again by colony-PCR. Only the colonies Nr. 2, 8 and 10 (see above) were checked (2 colonies each).
- Another colony-PCR of DH5a_pXy19_Ara
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- A5.13m_2 : 191.2 ng/µl 260/280= 1.84
- A5.13m_3 : 168.1 ng/µl 260/280= 1.78
- A2.5 : 193.0 ng/µl 260/280= 1.83
- A3.1m_5 : 146.1 ng/µl 260/280= 1.82
- A3.1m_1 : 250.1 ng/µl 260/280= 1.84
- A4.2m_4 : 124.8 ng/µl 260/280= 1.85
- Restriction digest of A5.13m_2, A5.13m_3, A3.1m_1, A3.1m_5, A2.5, A4.2m_4 with EcoRI and SpeI. The restriction was done to enable the ligation of the mutated genes into pSB1C3. Afterwards DH5a was transformed with the ligation product.
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- C505_2_2 : 253.6 ng/µl 260/280= 1.60
- C505_2_3 : 274.7 ng/µl 260/280= 1.88
- C505_8_2 : 462.1 ng/µl 260/280= 1.38
- C505_8_3 : 149.1 ng/µl 260/280= 1.84
- C505_10_3 : 160.4 ng/µl 260/280= 1.98
- Restriction digest of the Miniprep products with EcoRI and EcoRI/SpeI. Afterwards the restriction products were analyzed by Agarose gel electrophoresis. Gel loading: uncut, cut once, cut twice
- C505_2_3 und C505_8_2 were selected for mutagenic PCR.
- Restriction digest of pSB1C3 with EcoRI and SpeI
- Restriction digest of pXylE with EcoRI and XbaI and of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] with EcoRI and SpeI. Dephosphorylation of pXylE. Ligation of pXylE and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara]
week 9 (16.-20.07.12)
- mutagenic PCR of C505_2_3 and C505_8_2. (Did not work)
- [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808003 tct_B162m] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808005 tct_B197m] with primer to add a RBS in front and behind the gene
- Restriction digest of the PCR products with EcoRI and SpeI for ligation into pSB1C3_5.13m (tphC)
- Adding a RBS to one site of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] using PCR with the reverse primer
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- C3.1m_1 : 278.9 ng/µl 260/280= 1.88
- C4.2m_4 : 214.2 ng/µl 260/280= 1.86
- 5.13m_3 : 256.3 ng/µl 260/280= 1.88
- C2.5_2 : 44.8 ng/µl 260/280= 2.00
- C5.13m_2 : 60.8 ng/µl 260/280= 1.89
week 10 (23.-27.07.12)
- Restriction digest of pSB1C3_5.13m_3_2 and pSB1C3_5.13m_3_3 with EcoRI and XbaI. Afterwards the products are ligated with RBS_tctB162m_RBS and RBS_tctB197_RBS each.
- Restriction digest of A3.1m_1 for ligation with Ara
- [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] of Ara_RBS and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] for ligation into pXylE and pSB1C3. Afterwards restriction digest with restriction enzymes: Ara_RBS is cut with with EcoRI and SpeI and Ara is cut with EcoRI and XbaI
- Ligation of the restricted PCR-products to: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE and transformation into DH5a
- mutagenic PCR of 505_8_2
week 11 (30.07.-03.08.12)
- Checking transformation of DH5a by colony-PCR: pSB1A2_tctB197_tphCm, pSB1C3_tctB162m_tphCm, pSB1C3_tct197_tphCm, pSB1A2_tctB162m_tphCm, pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_Ara_XylE. Positive colonies are picked and used for inoculation of fluid media.
- Miniprep (Fermentas Kit) of fluid culture and Nanodrop measurement of:
- pSB1C3_503 : 110.0 ng/µl 260/280= 1.82
- pSB1A2_tctB197_tphCm : 32.0 ng/µl 260/280= 2.10
- pSB1A2_Ara_tctA503m : 202.0 ng/µl 260/280= 1.78
- pSB1A2_tctB162m_tphCm : 36.5 ng/µl 260/280= 1.91
- pSB1C3_Ara_tctA503m : 133.2 ng/µl 260/280= 1.83
- pSB1C3_tctB162m_tphCm : 35.2 ng/µl 260/280= 1.93
- pSB1C3_tct197_tphC : 30.6 ng/µl 260/280= 1.88
- Restriction digest of pSB1A2_Ara_tctA503m, pSB1C3_Ara_tctA503m, pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm and building the final constructs by ligation: pSB1A2_tctA503m_tctB162m_tphCm und pSB1C3_tctA503m_tctB162m_tphCm. Transformation of the final constructs into DH5a.
- Transformation of pSB1C3_503 into DH5a
- Checking the mutagenic PCR product of 505_8_2 and the transformation of DH5a with pSB1C3_503 by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR]
- Restriction digest of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] und C4.2_4_1 using EcoRI and PstI, building pSB1C3_Ara by ligation and transformation of the vector into DH5a
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- pSB1C3_Ara : 86.7 ng/µl 260/280= 1.83
- pSB1C3_505m : 113.8 ng/µl 260/280= 1.85
- Restriction digest of pSB1C3_505m with PstI. Checking if the restriction site has been successfully removed
- restriction site hasn't been removed
- Screnning for transformation of DH5a with pSB1A2_tctA503m_tctB162m_tphCm and pSB1C3_tctA503m_tctB162m_tphCm using colony-PCR [*]
- Checking the constructs pSB1A2/pSB1C3_Ara_503m and pSB1A2/pSB1C3_tctB162m_tphCm by [http://en.wikipedia.org/wiki/Polymerase_chain_reaction PCR] [*]
- Ligation of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 Ara] into pSB1C3
- Checking if pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm have the right Prefix/Suffix by digesting both with EcoRI and XbaI and with EcoRI and PstI [*]
- Ligation of pSB1A2_tctB162m_tphCm and pSB1C3_tctB162m_tphCm with Ara_503
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- pSB1A2_tctB162m_tphCm_1 : 45.3 ng/µl 260/280= 1.90
- pSB1A2_tctB162m_tphCm_2 : 74.5 ng/µl 260/280= 1.82
- pSB1C3_tctB162m_tphCm_1 : 95.8 ng/µl 260/280= 1.86
- pSB1C3_tctB162m_tphCm_2 : 85.7 ng/µl 260/280= 1.81
- pSB1C3_tctB162m_tphCm_3 : 100.1 ng/µl 260/280= 1.84
- pSB1C3_tctB162m_tphCm_4 : 79.9 ng/µl 260/280= 1.88
- pSB1C3_tctB162m_tphCm_5 : 89.8 ng/µl 260/280= 1.85
- Sequencing pSB1A2_tctB162m_tphCm_2 and pSB1C3_tctB162m_tphCm_5
week 12 (06.-10.08.12)
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- pSB1A2_tctB162m_tphCm_2 : 55.7 ng/µl 260/280= 1.81
- pSB1C3_tctB162m_tphCm_5 : 101.2 ng/µl 260/280= 1.92
- Mutagenic PCR of pSB1C3_505_8_2 and subsequent transformation into DH5a
- Ligation of Ara_503m with pSB1A2_tctB162m_tphCm/pSB1C3_tctB162m_tphCm and subsequent transformation into DH5a. Screening the transformants using colony-PCR
- only colony 1, 2 and 3 have the right insert
- Miniprep (Fermentas Kit) and Nanodrop measurement of:
- pSB1A2_Ara_tctA503m : 278.0 ng/µl 260/280= 1.86
- pSB1C3_Ara_tctA503m : 169.0 ng/µl 260/280= 1.87
- pSB1C3_Ara_1 : 59.0 ng/µl 260/280= 1.90
- pSB1C3_Ara_4 : 55.5 ng/µl 260/280= 1.80
- pSB1C3_Ara_tctA503m_tctB162m_tphCm : 188.8 ng/µl 260/280= 1.86
- pSB1C3_Ara_tctA503m_tctB162m_tphCm :168.8 ng/µl 260/280= 1.84
- pSB1C3_Ara_tctA503m_tctB162m_tphCm :271.3 ng/µl 260/280= 1.85
- pSB1C3_Ara_XylE : 149.8 ng/µl 260/280= 1.86
- pSB1C3_tctB197_tphC : 95.8 ng/µl 260/280= 1.84
- pSB1A2_tctB197_tphC : 69.0 ng/µl 260/280= 1.88
- Sequencing of 3 different pSB1C3_Ara_tctA503m_tctB162m_tphC
- Analysis of mutagenic PCR products of 505_8_2 and restriction digest with PstI
- Restriction digest of Ara_RBS with EcoRI and SpeI and of pSB1C3_505m with EcoRI and XbaI. Building pSB1C3_Ara_505m by ligation and subsequent transformation into DH5a.
week 13 (13.-17.08.12)
- Restriction digest of pSB1C3_197_tphCm with EcoRI and XbaI
- Checking the transformation of DH5a with pSB1C3_Ara_505m by using colony-PCR
- Sequencing pSB1C3_Ara_XylE, pSB1C3_Ara_tctA503m_tctB162m_tphCm (=Operon1), pSB1C3_505m
- Restriction digest of Ara_RBS with EcoRI and SpeI and of pSB1C3_tphCm with EcoRI and XbaI. Building pSB1C3_Ara_tphCm by ligation and subsequent transformation into DH5a
- Restriction digest of pSB1C3_Ara_tctA505m with EcoRI and SpeI. Ligation of Ara_tctA505m and tctB197_tphCm into pSB1C3
week 14 (20.-24.08.12)
- Checking the transformation of DH5a with pSB1C3_Ara_tphCm using colony-PCR
week 15 (27.-31.08.12)
- Restriction digest of pSB1C3_Ara_tphCm and pSB1C3_Ara_tctA505m with PstI and SpeI
- Restriction digest of pSB1C3_197_tphCm with XbaI and PstI.
- Building the construct pSB1C3_Ara_tctA505m_197_tphCm (= Operon2) by ligation. Subsequent transformation into DH5a
- Checking the transformation of DH5a with Operon2 using colony-PCR
 "
"