Team:Bielefeld-Germany/Labjournal/week19
From 2012.igem.org
(Difference between revisions)
(→Tuesday September 4th) |
(→Wednesday September 5th) |
||
| Line 26: | Line 26: | ||
===Wednesday September 5th=== | ===Wednesday September 5th=== | ||
| + | * '''Team Cultivation & Purification:''' | ||
| + | ** Performed the cell disruption of yesterday's flask cultivation to produce CBD-GFP+His fusion protein via sonification and made a SDS-Page of it. The problem is to purify this product, because it might be impossible to remove the protein from the column. If it binds via CBD and not via His-tag. | ||
| + | ** Made a flask cultivation of ''E.coli'' KRX containing new BioBricks: Laccase from ''E.coli'', ''B.pumilus'' and ''T.thermophilus'' behind a constitutive promoter. -->Settings: 100 mL flask without baffles, autoinduction medium, final volume: 60 mL, 37 °C, 120 rpm, 12 hours, double determination. | ||
| + | ** We repeated the purification of His-tagged GFP by using the flow-through and the washing fraction, but it did not work. Then we got to know that it could not work, because the sequencing showed us that the GFP was not tagged to His. | ||
| + | ** Made a SDS-Page of the fractions of purified ''B.pumilus'' laccase produced at 09/03. | ||
| + | ** Starting of another fermentation of laccase from ''B.pumilus'' to get a growth kinetics. -->Settings: fermenter: Braun, autoinduction medium, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NF/m, 6 hours. Found out that we got a maximal optical density after 4 hours, after which the cells start to die. Therefore we harvested the cells after 6 hours. | ||
| + | |||
===Thursday September 6th=== | ===Thursday September 6th=== | ||
===Friday September 7th=== | ===Friday September 7th=== | ||
Revision as of 11:15, 18 September 2012
Contents |
Week 19 (09/03 - 09/09/12)
Monday September 3rd
- Team Cultivation & Purification:
- Made one half of the SDS-Pages of the flask cultivation from 08/30.
- Work on fermentation of B.pumilus laccase (08/31) was continued and we performed the cell disruption by high-pressure homogeniser in 3 cycles. Then it was purificated by using a TALON column. The product was given to the activity team for analysis.
- Fermentation of E.coli KRX with His-tagged GFP for the cellulose binding team --> Settings: fermenter: Braun, autoinduction medium, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NF/m. This time we cultivated only for 9 hours and then decreased the temperature at 20°C for a better folding of the protein for 1 hour. We had some problems with cascade at the end of the fermentation.
- Cells of today's fermentation of His-tagged GFP were disrupted via high-pressure homogeniser and laccase was purified by using the Talon column. The flow-through fluoresced very strong, so that not that much of the protein had bound to the column. Conclusion: The column had to be reloaded.
- Fermentation of E.coli KRX with laccase from B.pumilus. --> Settings: fermenter: Braun, autoinduction medium, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NF/m, 9 hours. We only cultivated 10 hours, because we saw that a great amount of our cells already died.
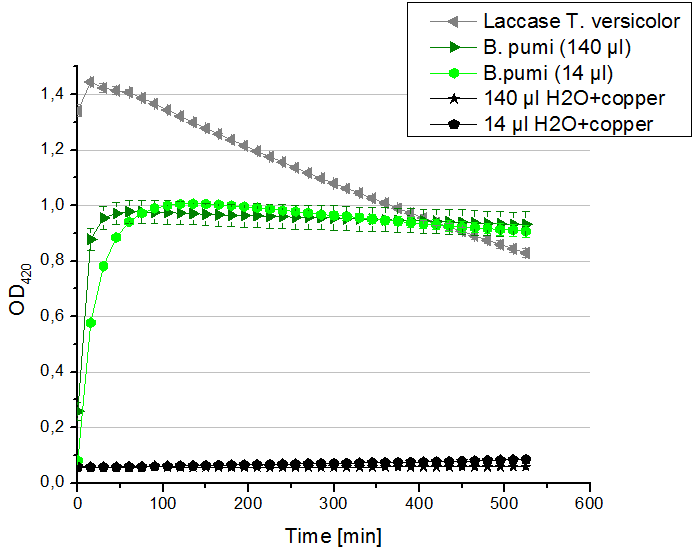
- Team Activity Tests: Today our hard-working Team Cultivation offered us some laccases from B. pumi. After fermentation and TALON purification the laccase was still hanging around in imidazole buffer, so we rebuffered it into nanopure water. We incubated the sample with copper and considered to measure the protein concentration before running the activity tests. Using the protocol for determining the protein concentration via Bradford (standards were 1 mg/ml, 1.5 mg/ml and 2 mg/ml BSA) we got a final B. pumi laccase concentration of 0.22 mg/ml. With this information we designed the following activity test different as usual. As a positive control we took 140 µl of our bought laccase from T. versicolor with a concentration of 0,021 mg/ml. Accordingly to this concentration we applied 14 µl of the B. pumi laccase to get approximately the same amount of laccase into each well. Additionally we tested 140 µl of the B. pumi laccase to make sure that we will definitely see any activity. After measuring the samples the whole night we got our results in the morning. It turned out, that our store bought laccase was rapidly active as seen before in our activity tests. The B. pumi laccase was also active. Using 140 µl of it led to the maximal state of oxidation of ABTS after 45 minutes. With the reduced amount of 14 µl of laccase this optimum was reached after 90 minutes. For our following activity tests we want to include the protein concentration again.
Tuesday September 4th
- We signed in for our tracks today! Our first choice is the 'Environment' track since it is obvious that we want to clean up drinking water and therefore help our environment. The second choice is the 'New Application' track. We are hoping our filter system can be applied in wastewater treatment plants and establishes a new area of water purification.
- Team Cultivation & Purification:
- Second half of the SDS-Pages of the flask cultivation from 08/30 was made.
- We reloaded our TALON column.
- After reloading we repeated the purification of B.pumilus laccase produced by fermentation on 08/31. Therefore the flowthrough and the washing fraction were used. The different fractions should be tested by the activity team.
- A new fermentation of E.coli KRX with E.coli laccase was started --> Settings: fermenter: Infors, autoinduction medium, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NF/m, 9 hours. We shorten the time, because the effect should be similar to the production of B.pumilus laccase.
- The fermentation of E.coli KRX with B.pumilus laccase and also with E.coli laccase were harvested and after centrifugation stored at 4°C, because our high-pressure homogeniser is out of work.
- Made a flask cultivation for the cellulose binding team: cultivation of E.coli KRX containing a plasmid with cellulose binding domain (CBD) fused to His-tagged GFP.-->Settings: 1 L flask with baffles, autoinduction medium, final volume: 250 mL, 37 °C, 120 rpm, 9 hours. We made a three-fold determination, but did not get fluorescing cultures. The problem is to purify this product, because it might be impossible to remove the protein from the column. If it binds via CBD and not via His-tag.
Wednesday September 5th
- Team Cultivation & Purification:
- Performed the cell disruption of yesterday's flask cultivation to produce CBD-GFP+His fusion protein via sonification and made a SDS-Page of it. The problem is to purify this product, because it might be impossible to remove the protein from the column. If it binds via CBD and not via His-tag.
- Made a flask cultivation of E.coli KRX containing new BioBricks: Laccase from E.coli, B.pumilus and T.thermophilus behind a constitutive promoter. -->Settings: 100 mL flask without baffles, autoinduction medium, final volume: 60 mL, 37 °C, 120 rpm, 12 hours, double determination.
- We repeated the purification of His-tagged GFP by using the flow-through and the washing fraction, but it did not work. Then we got to know that it could not work, because the sequencing showed us that the GFP was not tagged to His.
- Made a SDS-Page of the fractions of purified B.pumilus laccase produced at 09/03.
- Starting of another fermentation of laccase from B.pumilus to get a growth kinetics. -->Settings: fermenter: Braun, autoinduction medium, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NF/m, 6 hours. Found out that we got a maximal optical density after 4 hours, after which the cells start to die. Therefore we harvested the cells after 6 hours.
Thursday September 6th
Friday September 7th
Saturday September 8th
Sunday September 9th
Sunday
| 55px | | | | | | | | | | |
 "
"