Team:Bielefeld-Germany/Results/substrate
From 2012.igem.org
(→Degradation results) |
(→Degradation results) |
||
| Line 67: | Line 67: | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
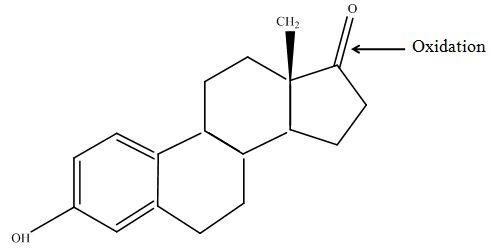
The Massspectrometry-Massspectrometry results showed us interesting peaks. Compare to the native estradiol component A shows peaks with 2 H atoms difference respectively component C for ethinly estradiol. Since we wanted to know more about our products we went to Prof. Dr. Dietmar Kuck, an organic chemister, to discuss our results. He suggest us two modells for the producs. First of them was that our laccases builds ketone reacting with the hydroxy group not in the phenolic ring. But this idea was incorrect because we could see 2 H less molecule also in ethinyl estradiol and the ethin group could not react with oxygen (Fig 17). Our next idea was, as described in the literature, that our laccase dehydrates the hydroxy group on the phenolic compond of the estradiol and ethinyl estradiol. This would make sence because it would build a chinon which you can see in Figure 18. | The Massspectrometry-Massspectrometry results showed us interesting peaks. Compare to the native estradiol component A shows peaks with 2 H atoms difference respectively component C for ethinly estradiol. Since we wanted to know more about our products we went to Prof. Dr. Dietmar Kuck, an organic chemister, to discuss our results. He suggest us two modells for the producs. First of them was that our laccases builds ketone reacting with the hydroxy group not in the phenolic ring. But this idea was incorrect because we could see 2 H less molecule also in ethinyl estradiol and the ethin group could not react with oxygen (Fig 17). Our next idea was, as described in the literature, that our laccase dehydrates the hydroxy group on the phenolic compond of the estradiol and ethinyl estradiol. This would make sence because it would build a chinon which you can see in Figure 18. | ||
| - | [[File:Bielefeld2012-estradiol-MS-kuck-suggest.JPG|thumb|350px|left|'''Figure 17: Chemical structure of the ketone after laccase treatment.''' This structure was designed with ChemBioDraw Ultra 12.0 after discussion with Mr. Kuck | + | [[File:Bielefeld2012-estradiol-MS-kuck-suggest.JPG|thumb|350px|left|'''Figure 17: Chemical structure of the ketone after laccase treatment.''' This structure was designed with ChemBioDraw Ultra 12.0 after discussion with Mr. Kuck on our MS/MS data.]] |
| - | [[File:Bielefeld2012-Estradiol-MSMS-possibleDegProduct.JPG|thumb|350px|right|'''Figure 18: Suggested Chemical structure of the oxydised estradiol.''' This structure was designed with ChemBioDraw Ultra 12.0 after discussion with Mr. Kuck | + | [[File:Bielefeld2012-Estradiol-MSMS-possibleDegProduct.JPG|thumb|350px|right|'''Figure 18: Suggested Chemical structure of the oxydised estradiol.''' This structure was designed with ChemBioDraw Ultra 12.0 after discussion with Mr. Kuck on our MS/MS data.]] |
<br style="clear: both" /> | <br style="clear: both" /> | ||
Revision as of 15:58, 26 October 2012

Contents |
Introduction
For analysis of degradation of different substrates some substrate analysis were made. For the measurements the four produced bacterial laccases (BHAL, ECOL, TTHL and BPUL) were used. The reactions were measured before and after a incubation time via high performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry. The HPLC was used particularly for analysis of degradation rates after defined time points. With this results it is possible to compare the different laccases in respect to their degradation feasibilities. To detect degradation products of estradiol and ethinyl estradiol after laccase treatment different analysis via LCMS and LCMS-MS were done. We identified two componds for each estradiol and ethinyl estradiol which are related to be products after laccase treatment.
Degradation measurements with high performance liquid chromatography
Dilution series of different estrogens
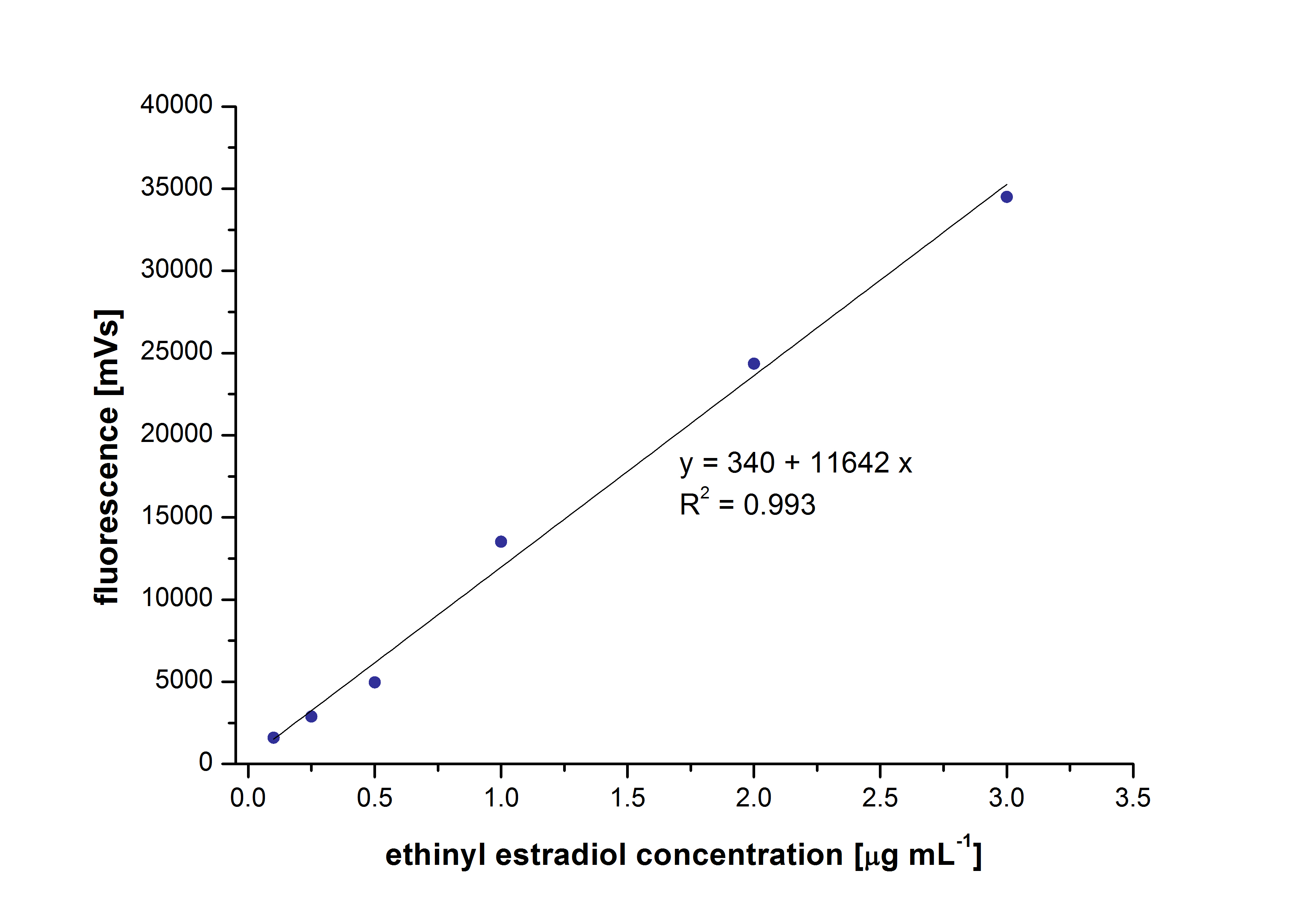
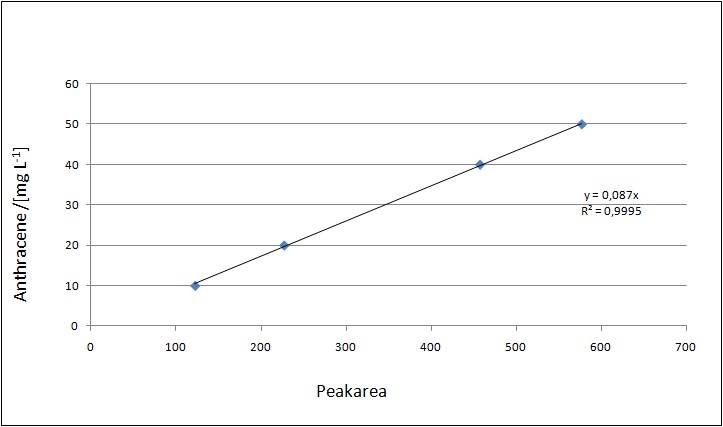
At first dilution series of all different substrates were measured. It was possible measure estradiol and ethinyl estradiol calibration curves but not for estrone. This was probably caused by its bad solubility.
The retention time for estradiol is 4.4 minutes and for ethinyl estradiol 4.9 minutes. For all estrogens the same extinction and emission values could be used: Ex 230, Em310.
Degradation of estrogens
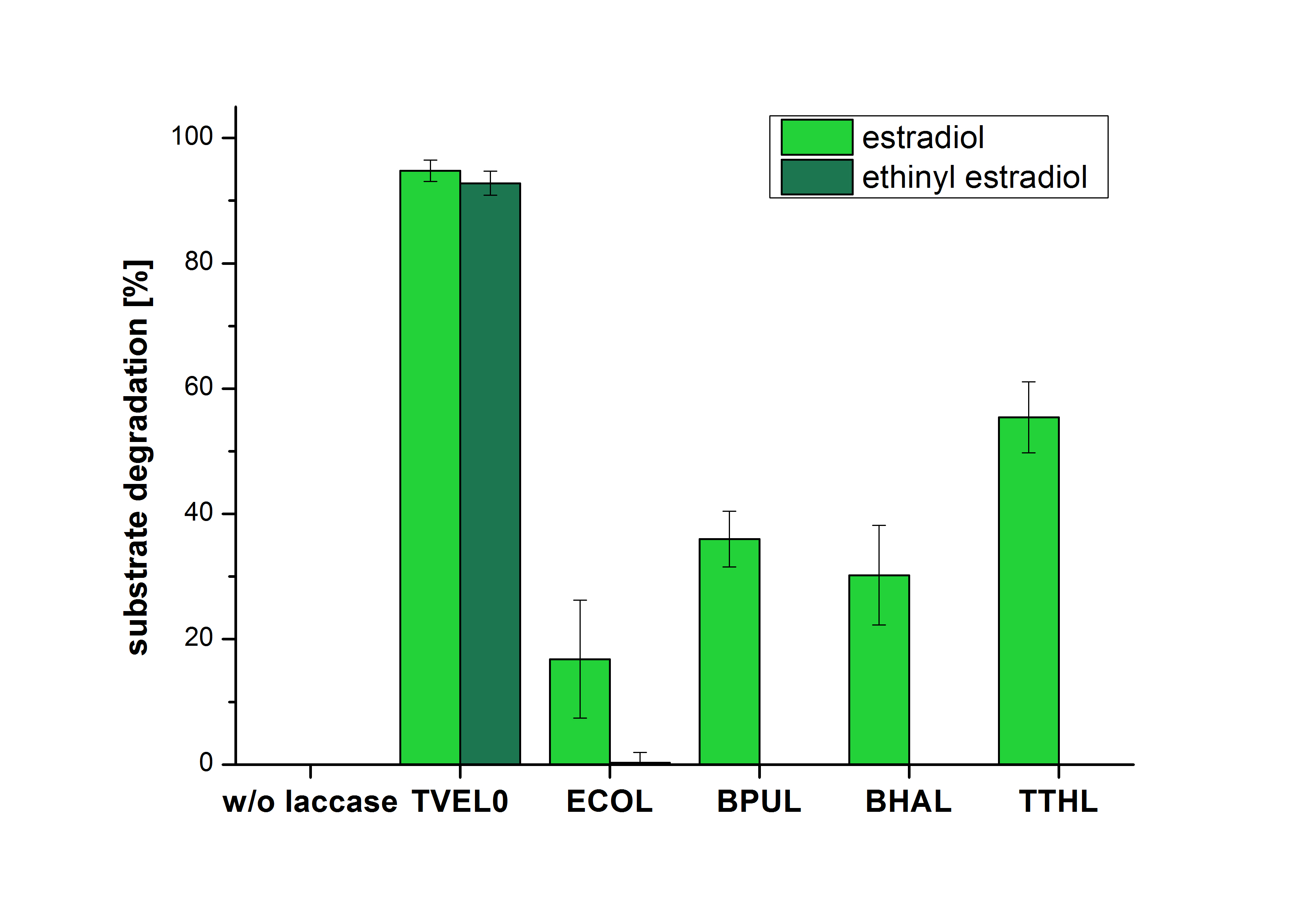
The measurements were made to test if the produced laccases were able to degrade different hormones. Therefore the produced laccases were inserted in the same concentrations (3 µg mL-1) to the different measurement approaches. For the correct pH value (which were measured by the Team Activity Test) Britton Robinson buffer at pH 5 was used for all measurements. The initial substrate concentration was 5 µg mL-1. The results of the reactions without ABTS are shown in Figure 1. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are indicated. The X-axis displays the different tested laccases. The degradation was measured at t0 and after five hours of incubation at 30 °C. The negative control was the substrate in Britton Robinson buffer and showed no degradation of the substrates. The bought laccase TVEL0 which is used as positive control is able to degrade 94.7 % estradiol and 92.7 % ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 35.9 % of used estradiol after five hours. ECOL was able to degrade 16.8 % estradiol. BHAL degraded 30.2 % estradiol. The best results were determined with TTHL (laccase from Thermus thermophilus). Here the percentage of degradation amounted 55.4 %.
The results of the reactions of the laccaes with addition of ABTS are shown in Figure 2. The experimental set ups were the same as the reaction approach without ABTS described above. The X-axis displays the different tested laccases. On the Y-axis the percentages of degraded estradiol (blue) and ethinyl estradiol (red) are shown. The degradation was measured at t0 and after five hours of incubation at 20 °C. The negative control showed no degradation of estradiol. 6.8 % of ethinyl estradiol were decayed. The positive control TVEL0 is able to degrade 100 % estradiol and ethinyl estradiol. The laccase BPUL (from Bacillus pumilus) degraded 46.9 % of used estradiol after ten minutes incubation. ECOL was able to degrade 6.7 % estradiol. BHAL degraded 46.9 % estradiol. With TTHL (laccase from Thermus thermophilus)a degradation 29.5 % were determined.

Spectrofluorophotometer Analysis
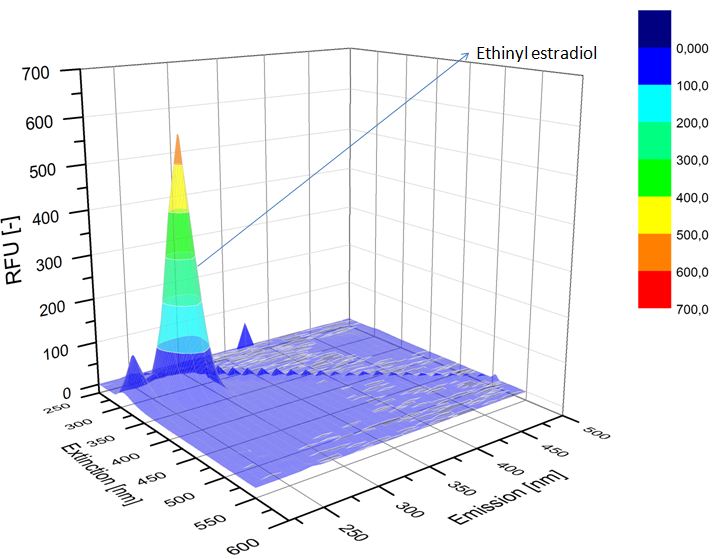
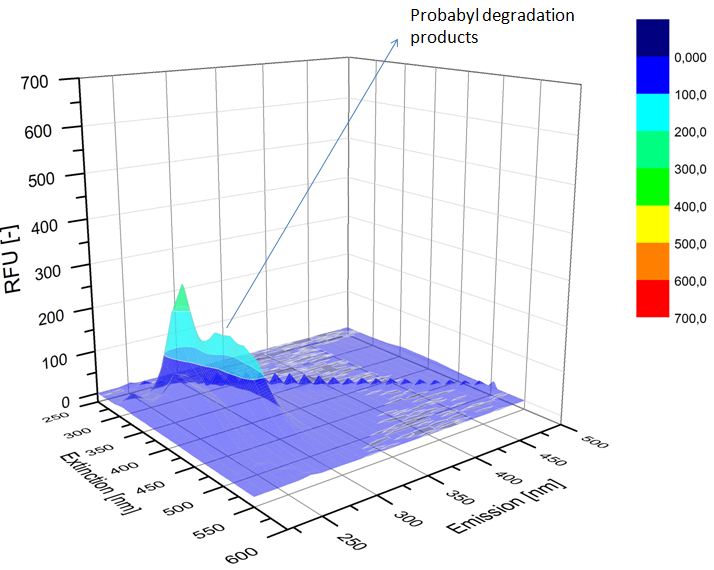
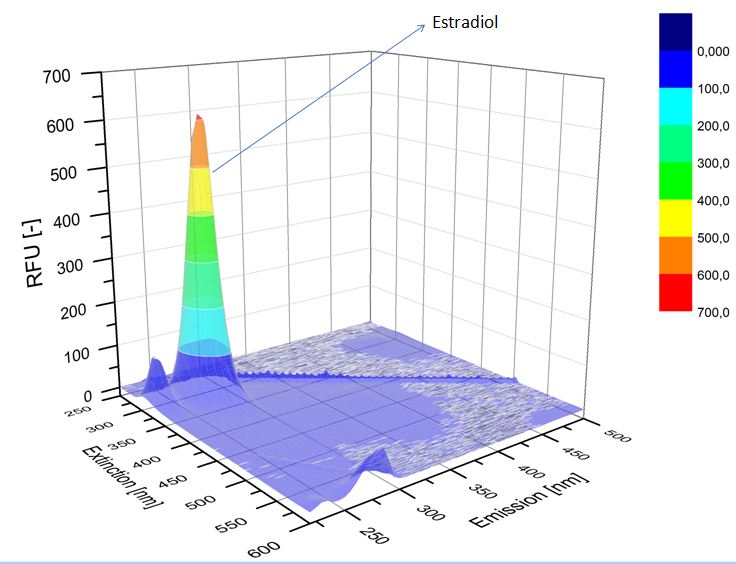
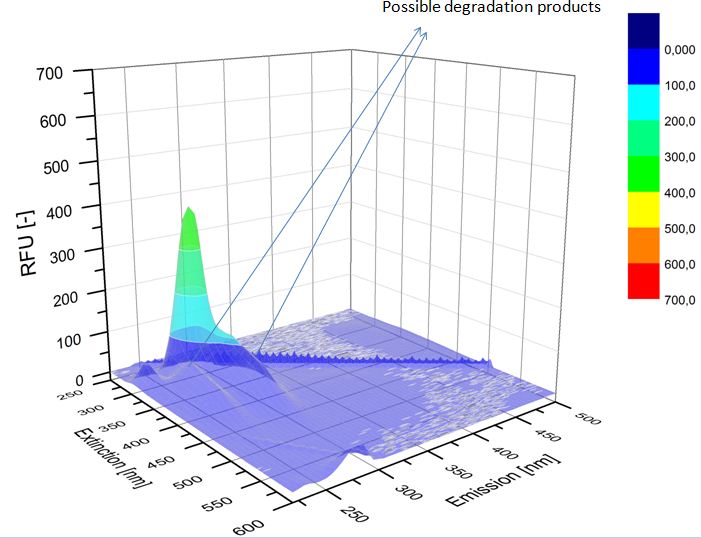
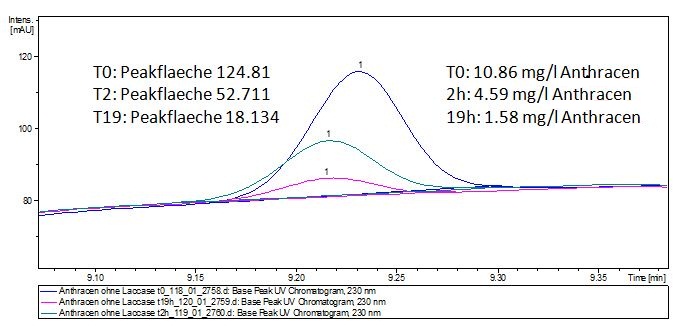
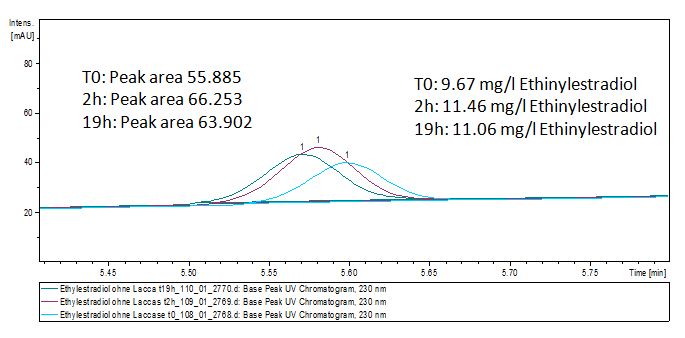
We analyzed the degradation of our substrates with the spectrofluorophotometer. As you can see it in the figures below the ethinyl estradiol and estradiol are degraded over night. Figure 3 shows the ethinyl estradiol without laccase treatment, Figure 4 shows that no more ethinyl estradiol can be detected in the sample after the degradation and new peaks appear, which might be represent possibly degradation products. In Figure 5 you can see the estradiol control without laccases. Like ethinyl estradiol our estradiol peak does not appear after the degradation and new peaks appear indicating that those are new degradation products.
Liquid chromatography–mass spectrometry
Dilution series
Our substrates are soluble in methanol. We set the standards to a concentration of 1 mg mL-1. The upper detection limit for the LC-MS was evaluated at concentration of 10 µg L-1 for the substrates estrone and estradiol. The same limit of detection was used for ethinyl estradiol and anthracene. We only used those four substrates. For all LC-MS sample preparations we used the T. versicolor laccases. The dilution series was prepared in methanol and 50 % acetonitril-water (v/v).
Degradation results
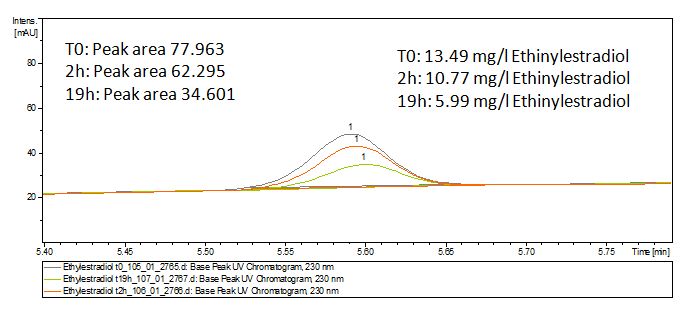
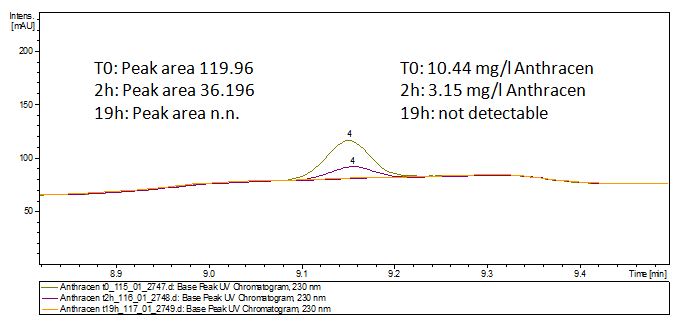
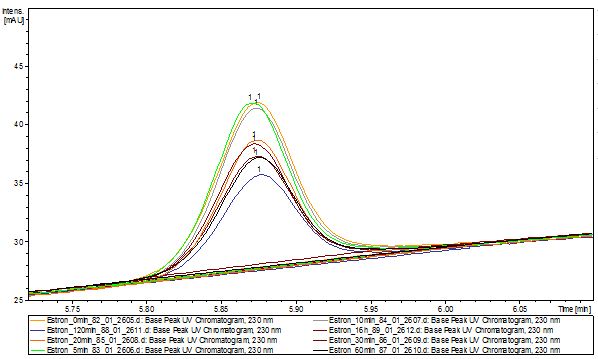
The TVEL0 was able to degrade the synthetic estradiol (Fig. 9) and probably anthracene (Fig. 11). The ethinyl estradiol control showed that it is stable in the used media (Fig. 10). Anthracene disintegrates in the Britton Puffer. But it could be observed, that there is less anthracene measurable with the LC-MS. The results indicate, that the laccase is able to degrade anthracene (Fig. 12). Estrone (Fig. 13) and estradiol (Fig. 14) were degraded as well. Using estrone it could not be identify any degradation products. The reason for this could be that the products are not detectable with LC-MS or with the applied methods. Peaks in the degradation of estradiol have been shown but we were not able to identify them. It could be degradation products. In the following figures the results of the LC-MS measurements are presented.
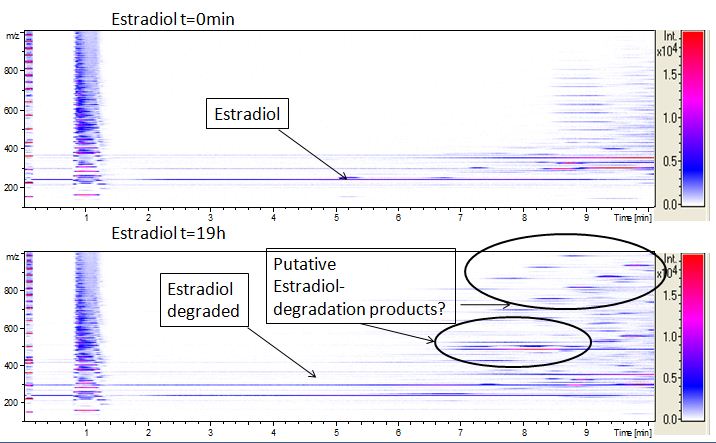
We also tried to measure the degradation using mass-spectrometry. Since quantification via mass-spectrometry is difficult regarding the ionization of the analytes, we quantified our substrates by UV-light. Nevertheless, mass spectrometry enables identification of possible degradation products. We analyzed estradiol degradation in detail (Fig. 14), resulting in the detection of possible chemical compounds generated during the (enzymatic) degradation. Until now we are not sure how estradiol degradation works, but with more time granted, the degradation products can be identified.
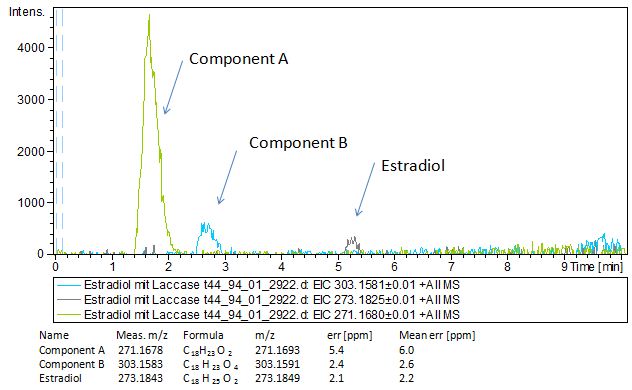
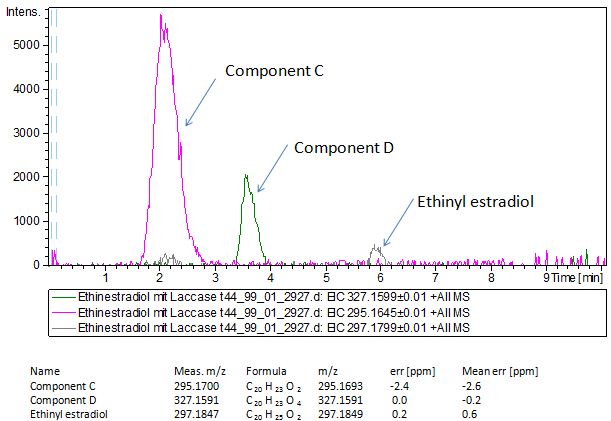
Since we have seen some possible degradation products, we used more estradiol, ethinyl estradiol and more laccase for the reaction to the LC-MS measurement. We found out that after the laccase treatment two new peaks appears in both estradiol and ethinyl estradiol. A check against databases could not identify those components so we did an MS-MS on component A. Since component B is fewer in concentration then component A, we could not find out anything about it (Fig. 15 and Fig. 16). To note the chemical formulares are M+H values. For the real chemical formular you have to deduct this H.
The Massspectrometry-Massspectrometry results showed us interesting peaks. Compare to the native estradiol component A shows peaks with 2 H atoms difference respectively component C for ethinly estradiol. Since we wanted to know more about our products we went to Prof. Dr. Dietmar Kuck, an organic chemister, to discuss our results. He suggest us two modells for the producs. First of them was that our laccases builds ketone reacting with the hydroxy group not in the phenolic ring. But this idea was incorrect because we could see 2 H less molecule also in ethinyl estradiol and the ethin group could not react with oxygen (Fig 17). Our next idea was, as described in the literature, that our laccase dehydrates the hydroxy group on the phenolic compond of the estradiol and ethinyl estradiol. This would make sence because it would build a chinon which you can see in Figure 18.
Further substrate analysis via HPLC
HPLC analysis of polycylic aromatic hydrocarbons
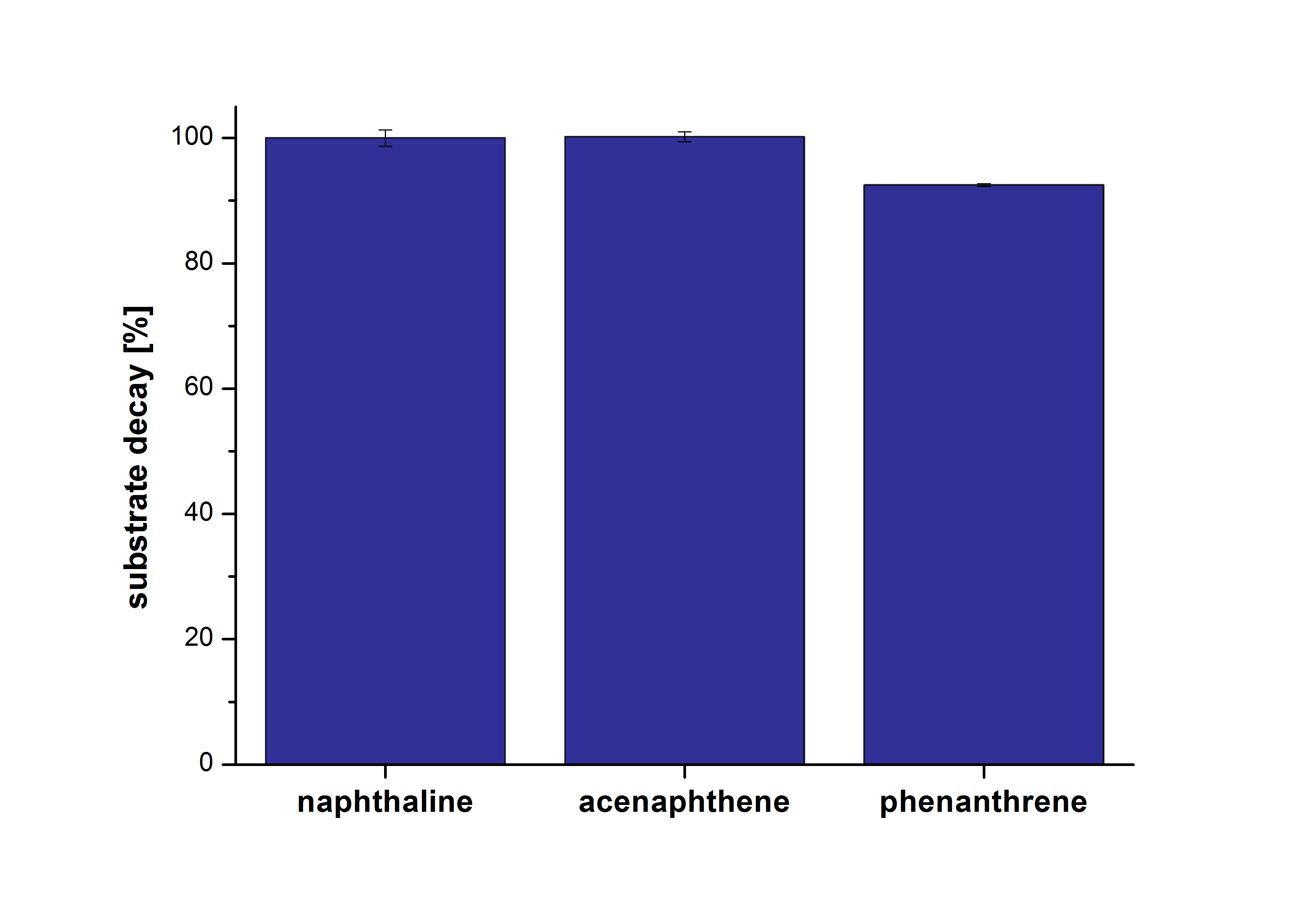
The results of our negative control measurements of polycyclic aromatic hydrocarbons showed that the PAHs decayed without laccase treatment after one hour in Britton Robinson (BR)-buffer. This is shown in Figure X.
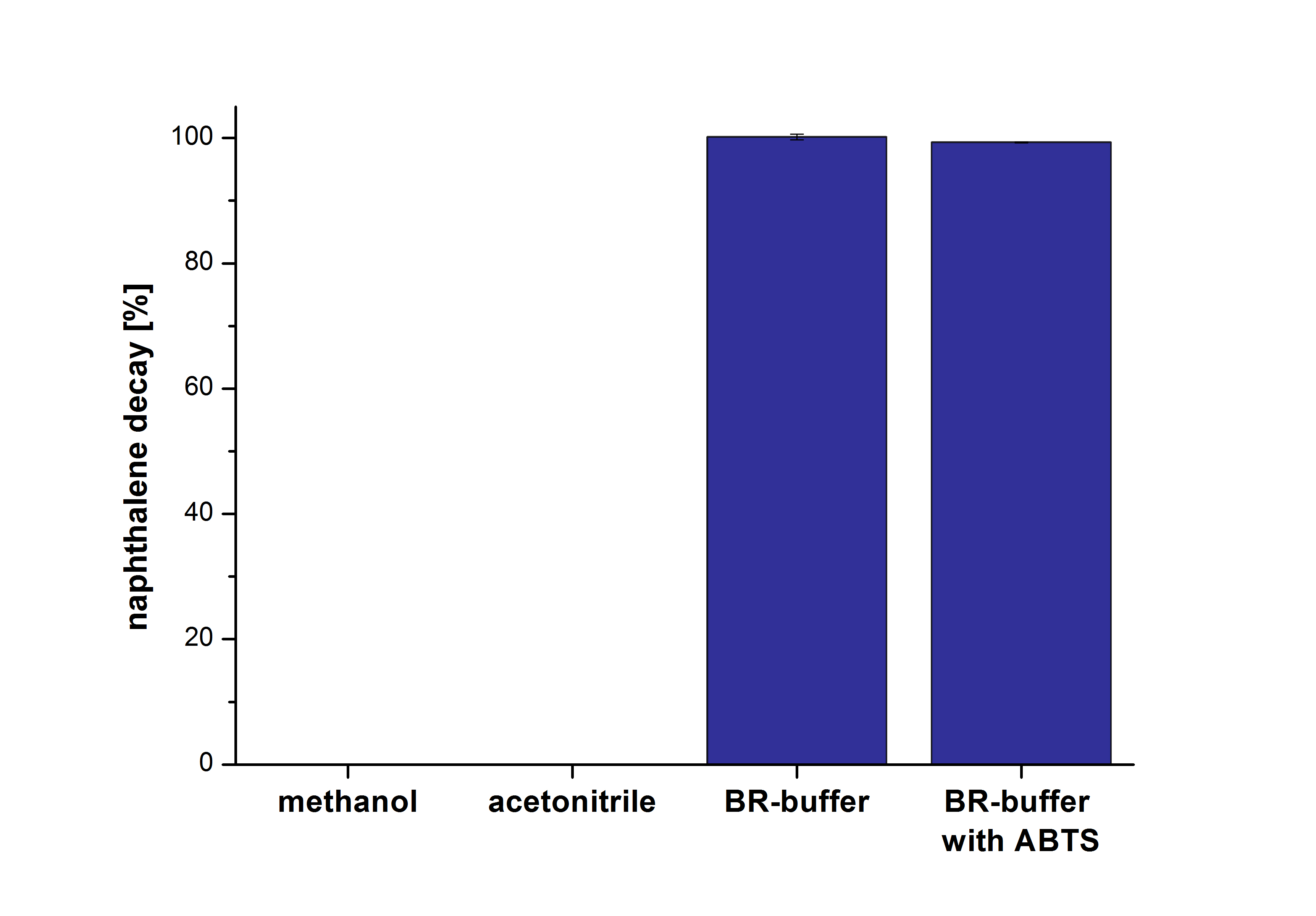
The next step was to check which substances in the reaction approach may cause the decay. Therefore naphthalene was dissolved in methanol (which is the solvent for the substrate and used for stopping the reaction), acetonitrile (which could be used as alternative solvent) and BR-buffer with and without ABTS. The results are shown in Figure Y. Using pure methanol and acetonitrile naphthalene is not decayed. In BR-buffer with and without ABTS the decay is nearly completed after one hour treatment. So BR-buffer seems a bad choice to test the degradation of naphtalene under laccase treatment.
HPLC analysis of analgesics
Another class of substrates were analgesics. For the three analgesics substrates we used we have different optimal extinction and emission values. Additional, difficulties occurred with ibuprofen. Instead of one single peak we found two and they didn't correlate with the used concentrations. With diclofenac the extinction and emission values were not found. Therefore the substrate was analyzed with the spectrofluorophotometer but this has also shown no clear peak for diclofenac and made it therefore not measurable.
end
| 55px | | | | | | | | | | |
 "
"