Team:Bielefeld-Germany/Protocols/Production
From 2012.igem.org
(→Sonification) |
|||
| Line 5: | Line 5: | ||
</html> | </html> | ||
| - | + | ||
=Production= | =Production= | ||
| - | + | ||
Here all our methods according to cultivation and purification are listed. | Here all our methods according to cultivation and purification are listed. | ||
| - | + | ||
==Pre-Cultivation== | ==Pre-Cultivation== | ||
| - | + | ||
===Precultivation of ''E.Coli'' KRX (with or without BioBrick)=== | ===Precultivation of ''E.Coli'' KRX (with or without BioBrick)=== | ||
* 50 mL [https://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB]-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs | * 50 mL [https://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB]-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs | ||
| Line 38: | Line 38: | ||
* Shaking at 140 rpm | * Shaking at 140 rpm | ||
| - | + | ||
==Cultivation== | ==Cultivation== | ||
| - | + | ||
===Expression of Laccase=== | ===Expression of Laccase=== | ||
| Line 71: | Line 71: | ||
* Harvest after 12-13 h | * Harvest after 12-13 h | ||
| - | + | ||
==Cell Harvesting== | ==Cell Harvesting== | ||
| - | + | ||
* Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C | * Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C | ||
| Line 79: | Line 79: | ||
* Resuspend the pellet in 5 mL [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#special_buffers_for_resuspending_the_cell_pellet special buffer] or [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography binding buffer] for each gramm of cell paste | * Resuspend the pellet in 5 mL [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#special_buffers_for_resuspending_the_cell_pellet special buffer] or [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography binding buffer] for each gramm of cell paste | ||
| - | + | ||
==Solubilization of inclusion bodies== | ==Solubilization of inclusion bodies== | ||
| - | + | ||
* centrifugation of the celllysate at 40,000 g for 30 minutes | * centrifugation of the celllysate at 40,000 g for 30 minutes | ||
| Line 89: | Line 89: | ||
* resuspend the pellet in SDS running buffer | * resuspend the pellet in SDS running buffer | ||
| - | + | ||
=Cell disruption strategies= | =Cell disruption strategies= | ||
| - | + | ||
==B-PER lysis (chemical lysis)== | ==B-PER lysis (chemical lysis)== | ||
| Line 126: | Line 126: | ||
The lysate is ready for a following purification step. | The lysate is ready for a following purification step. | ||
| - | + | ||
==Mechanical lysis== | ==Mechanical lysis== | ||
| - | + | ||
The method of choice to disrupt the cells depends on the amount of biomass. | The method of choice to disrupt the cells depends on the amount of biomass. | ||
| - | + | ||
===Mechanical lysis of the (shaking flask) cultivation=== | ===Mechanical lysis of the (shaking flask) cultivation=== | ||
| - | + | ||
====Sonication==== | ====Sonication==== | ||
| Line 147: | Line 147: | ||
**fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes) | **fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes) | ||
**homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles. | **homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles. | ||
| - | + | ||
===Mechanical lysis of the (bio-reactor) cultivation=== | ===Mechanical lysis of the (bio-reactor) cultivation=== | ||
| - | + | ||
====Cell disruption with a high-pressure homogenizer==== | ====Cell disruption with a high-pressure homogenizer==== | ||
| Line 156: | Line 156: | ||
* disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much | * disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much | ||
| - | + | ||
=Purification= | =Purification= | ||
| - | + | ||
==His-tag affinity chromatography== | ==His-tag affinity chromatography== | ||
Revision as of 00:05, 27 September 2012
Contents |
Production
Here all our methods according to cultivation and purification are listed.
Pre-Cultivation
Precultivation of E.Coli KRX (with or without BioBrick)
- 50 mL LB-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 37 °C
- Shaking at 140 rpm
Precultivation of E.Coli Rosetta Gami 2 (with or without BioBrick)
- 50 mL LB-medium with 60 mg L-1 chloramphenicol and with 300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 37 °C
- Shaking at 140 rpm
Precultivation of Pichia Pastoris GS115 (complex-medium)
- 50 mL YPD-medium in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 30 °C
- Shaking at 140 rpm
Precultivation of Pichia Pastoris GS115 (minimal-medium)
- 50 mL YNB-medium in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 30 °C
- Shaking at 140 rpm
Cultivation
Expression of Laccase
- Chassis: Promega's E.Coli KRX
- Medium: Autoinduction-Medium supplemented with Chloramphenicol (final concentration 60 μg mL-1)
Cultivation with E. coli KRX in shaking flask(with baffles):
- 200 mL culture in 1000 mL shaking flask with baffles (Schott) with silicon plugs
- Cultivation temperature: 37 °C
- Autoinduction-medium with 20-60 mg L-1 chloramphenicol and if nessassary with 100-300 mg L-1 ampicillin
- Shaking at 140 rpm
- for characterizations: automatic sampling every 30 min
Bioreactor cultivations with E. coli KRX
To obtain higher amounts and concentration of proteins we cultivated and expressed in a bioreactor. It is possible to cultivate several liters and to control temperature, pH and pO_2.
- Bioreactor: Braun Biostat B Bioreactor (3L), Infors Labfors Bioreactor (3L), Bioengineering NLF22 Bioreactor (7 L),
- Autoinduction-medium with 60 mg L-1 chloramphenicol
- Culture volume: 3,0-6,0 L
- Starting OD600: 0.1 - 0.2
- Airflow: 5 NL/min
- pO2-Control: 30 % airsaturation (controlled with stirrer cascade starting with 200 rpm)
- pO2=100% calibration with 300rpm
- pH: 7.0 (controlled with 2M phosphoric acid and 2 M NaOH)
- Antifoam: BASF pluronic PE-8100
- Harvest after 12-13 h
Cell Harvesting
- Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C
- if the purification should start the next day store the cell pellet at 4°C !(the laccase must not be frozen!)
- Resuspend the pellet in 5 mL special buffer or binding buffer for each gramm of cell paste
Solubilization of inclusion bodies
- centrifugation of the celllysate at 40,000 g for 30 minutes
- resuspend the pellet of the lysate in 1 mL 6M Urea solution, incubation for 1 hour
- centrifugation for 10 minutes at 10,000 rpm
- resuspend the pellet in SDS running buffer
Cell disruption strategies
B-PER lysis (chemical lysis)
B-Per bacterial Protein Extraction Reagnt was used for a cell disruption screening according to the following protocol of Thermo Scientific.
- add 4 mL B-Per Reagent per gram of cell pellet
- resuspend the cell pellet by pipetting the suspension up and down until it is homogenous.
- incubate the solution for 10- 15 min at room temperature
- centrifuge lysate at 15.000g for 10 min
- decant the supernatant in a clean tube
The lysate is ready for a following purification step.
enzymatical lysis with lysozym
The lysis with lysozym was used for a cell disruption screening. The following protocol was utilized:
- resuspend the cell pellet in 600 µL of lysozym-solution per gramm
- incubate the solution for 1 h at 4°C
- centrifuge the lysate toseperate soluble proteins from insoluble proteins and cell debris.
combination of chemical and enzymatical lysis
B-Per bacterial Protein Extraction Reagnt was used for a cell disruption screening according to the following protocol of Thermo Scientific.
- add 4 mL B-Per Reagent per gram of cell pellet
- add 2µL of lysozym-solution(50 mg mL -1) and 2 µL DNaseI (2500 U mL -1) per mL B-Per Reagent. (For laccases:Do not use EDTA!)
- resuspend the cell pellet by pipetting the suspension up and down until it is homogenous.
- incubate the solution for 10- 15 min at room temperature
- centrifuge lysate at 15.000g for 10 min
- decant the supernatant in a clean tube
The lysate is ready for a following purification step.
Mechanical lysis
The method of choice to disrupt the cells depends on the amount of biomass.
Mechanical lysis of the (shaking flask) cultivation
Sonication
- Sonication of the re-suspended pellet on ice
- cycle number depends on the volume of the resuspended cells (e.g. 3 mL means 3 cycles)
- one cycle means sonification treatment for 1,5 min with Sonifier 450 by Branson, max. 50 %, cooled on ice, make sure not to heat the cells too much
Precellyse 24 homogenization
- homogenization with the Precellyse 24
- fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes)
- homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles.
Mechanical lysis of the (bio-reactor) cultivation
Cell disruption with a high-pressure homogenizer
- high-pressure homogenisation with a Rannie Homogenizer:
- disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much
Purification
His-tag affinity chromatography
- For buffers see here
Syringe method
- Column: 1 mL HisTrap FF crude by GE Healthcare
- Equilibrate with binding buffer(10mL)
- Load sample onto column(max. 6 mL)
- Wash with 10 mL binding buffer
- Elute with 5 mL of elution buffer
- Collect the eluate in 1 mL fractions, the purified protein is most likely in the first or second fraction
- Re-equilibrate the column with binding buffer
ÄKTA method
- Columns:
- 15 mL HisTrap FF crude by GE Healthcare
- 50 mL TALON-Histag-Purification Resin by Clonetech
Column preparation
- If Column is not loaded with Ni-ions /Cobalt-ions:
- Wash column with 5 - 8 Columnvolumes (CV) of deionized water
- Load column with metal-ions(4 CV)
- For HisTrap FF crude: 1,4% NiSO4-Solution
- For TALON-Histag-Purification Resin: CoCl2-Solution
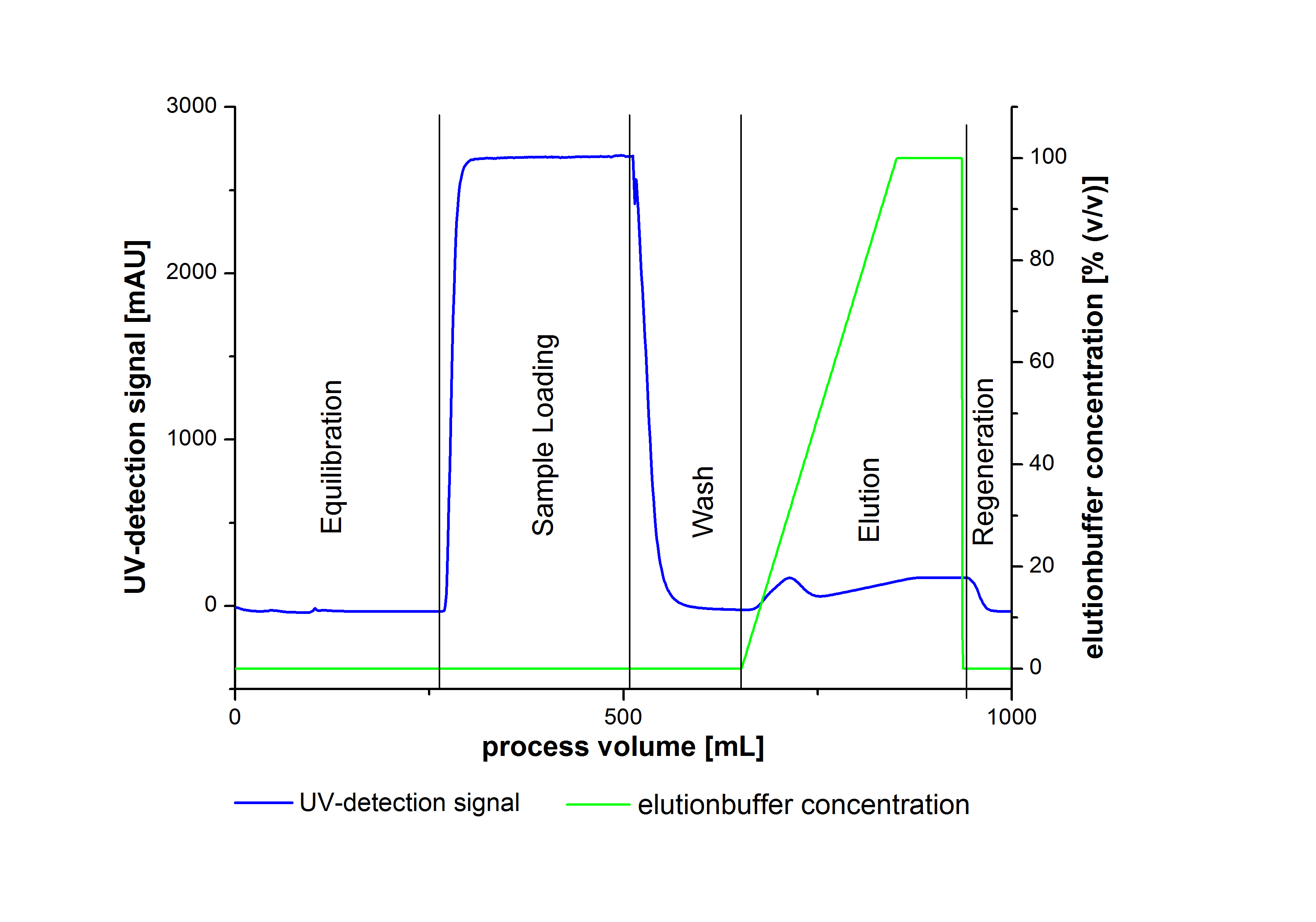
Chromatography protocol for the Äkta-system
- Wash column with 10 CV of deionized water
- Equilibrate column with 10 CV of binding buffer
- Load column with supernatant of the lysed cells (Collect the Flow through for SDS-PAGE analysis)
- Wash Column with 10 CV of binding buffer (Collect the Flow through for SDS-PAGE analysis)
- Elute Protein with an increasing elutionbuffer ratio (gradient 0%-100%, length 200mL)
- Collect the eluate in 10 mL fractions
- Elute remaining proteins with 100% Elutionbuffer (4 CV)
An typical chromatogram of purified laccases is illustrated in the following grafic:
| 55px | | | | | | | | | | |
 "
"