Team:NTU-Taida/Modeling/System-Analysis
From 2012.igem.org
| Line 11: | Line 11: | ||
===System Robustness=== | ===System Robustness=== | ||
| - | *Protein Production Rate | + | *'''Protein Production Rate''' |
Revision as of 19:24, 26 September 2012
System Analysis
Contents |
Overview
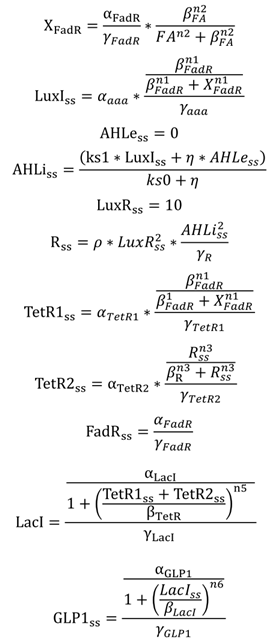
System analysis explores extensively into the parameter space by sweeping each parameters for certain range while make the others constant. In this way, we can see the corresponding output response when the value of a specific parameter varies within a certain extent. It provides deep insights into the roles each parameter plays in our single cell model. In one aspect, it examines the robustness of our system. In another aspect, it provides valuable information helping us to determine which parameters should be adjusted in order to achieve our expected circuit response, that is, to tune the threshold of our high-pass filter to the desired level.
Parameter Sweeping
Our system analysis model is an extension of our single cell model. In order to perform the parameter sweeping, we replace the constant values of parameters with a range of values. For simplicity, we focus only on the steady state of our circuit. The steady states of each species are derived using Maple, and are then ported to Matlab for simulation. The input of our system, the fatty acid, is also included as our sweeping parameters in each sweep, enabling us to observe the threshold of the filter in our circuit. The combined input and parameter sweeping gives us a 3-dimensional graph, with x-axis, y-axis and z-axis being the parameter under analysis, the input fatty acid concentration and the output GLP1 concentration, respectively.
Results
The following figures show the results of the sweeping of protein production rates, repression coefficients and degradation rates.
System Robustness
- Protein Production Rate
 "
"