Team:Paris Bettencourt/SID
From 2012.igem.org
Zmarinkovic (Talk | contribs) (→Overview) |
Zmarinkovic (Talk | contribs) (→Design) |
||
| Line 28: | Line 28: | ||
==Design== | ==Design== | ||
| - | |||
| - | + | The general design of the Synthetic Import Domain system is very simple. We take the binding and translocation domain from the wild-type colicin and clone it on a plasmid as "Synthetic Import Domain". After, we can fuse any protein of our choice and test if it is going to be imported into other E. coli cells. | |
| - | + | ||
| - | + | ||
[[File:Bettencourt_Sid3.png|frameless|center|800px]] | [[File:Bettencourt_Sid3.png|frameless|center|800px]] | ||
Revision as of 11:14, 25 September 2012
Contents |
Overview
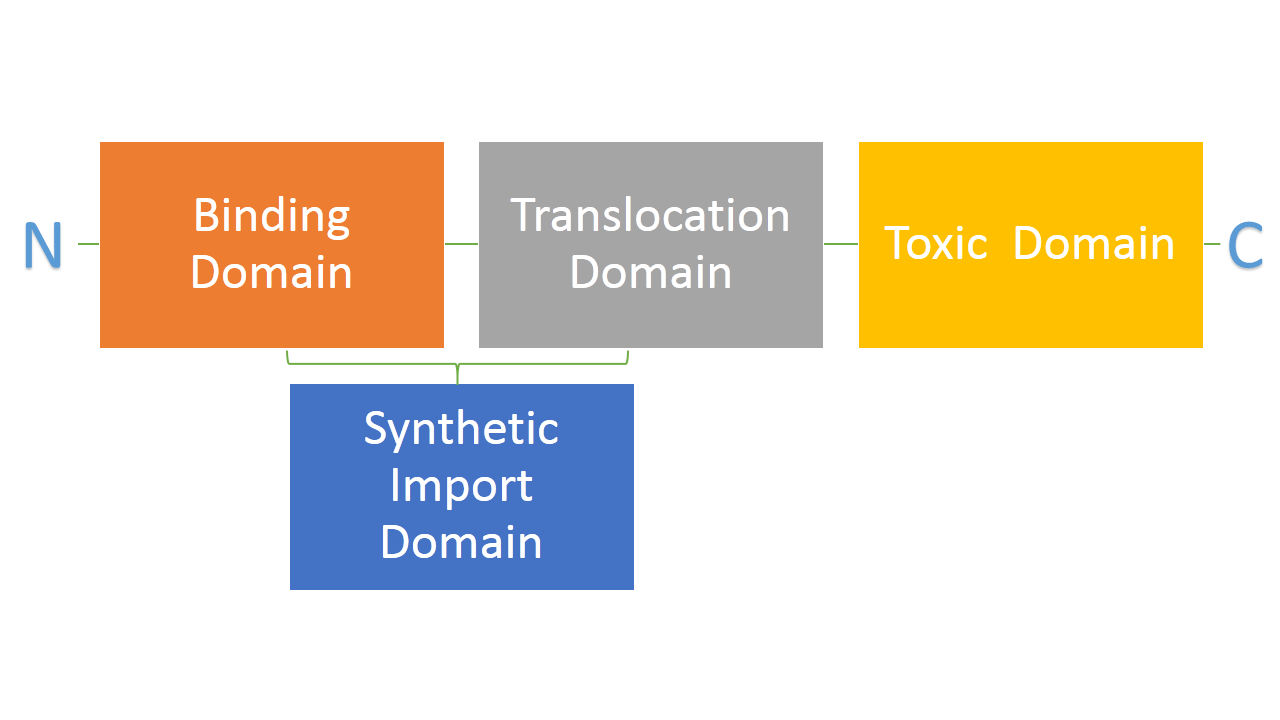
Bacteria developed mechanisms to kill other bacteria in order to reduce competition between themselves in the environment [1]. Some strains of E. coli produce lethal proteins called colicins which kill other bacteria, including E. coli. Colicins are built out of three main domains which are also a point of difference among many types of colicins. First domain is responsible for binding to a receptor on a bacterial membrane, second one is responsible for translocating the protein from outside to inside of a bacteria and the third one is responsible for killing bacteria. We are interested in two types of colicins, colicin E2 and colicin D. Both of them use different binding, translocation and killing mechanisms. Colicin E2 is a DNAse and colicin D is an RNAse.
We hypothesize that it is possible to use binding and translocation part of colicin as a "synthetic import domain" onto which we can fuse other proteins in order to import them into bacteria. This would permit us to create new communication system between E. coli cells through direct transfer of proteins form one cell to another. Such a system would have a significant impact in biological research and it would be an important addition to tools made by and used by synthetic biology. Great variance in possible binding and translocation domains we could use is also a big plus.
Main aim of this part of the project is to fuse the RNAse domain from colicin D to the "synthetic import domain" of colicin E2. By doing this we will obtain a toxin that targets the same receptor and translocation mechanism as colicin E2 but kills bacteria on the level of protein synthesis by cleaving tRNA as the RNAse domain of colicin D. This new toxin could be used in our project as an additional Suicide System.
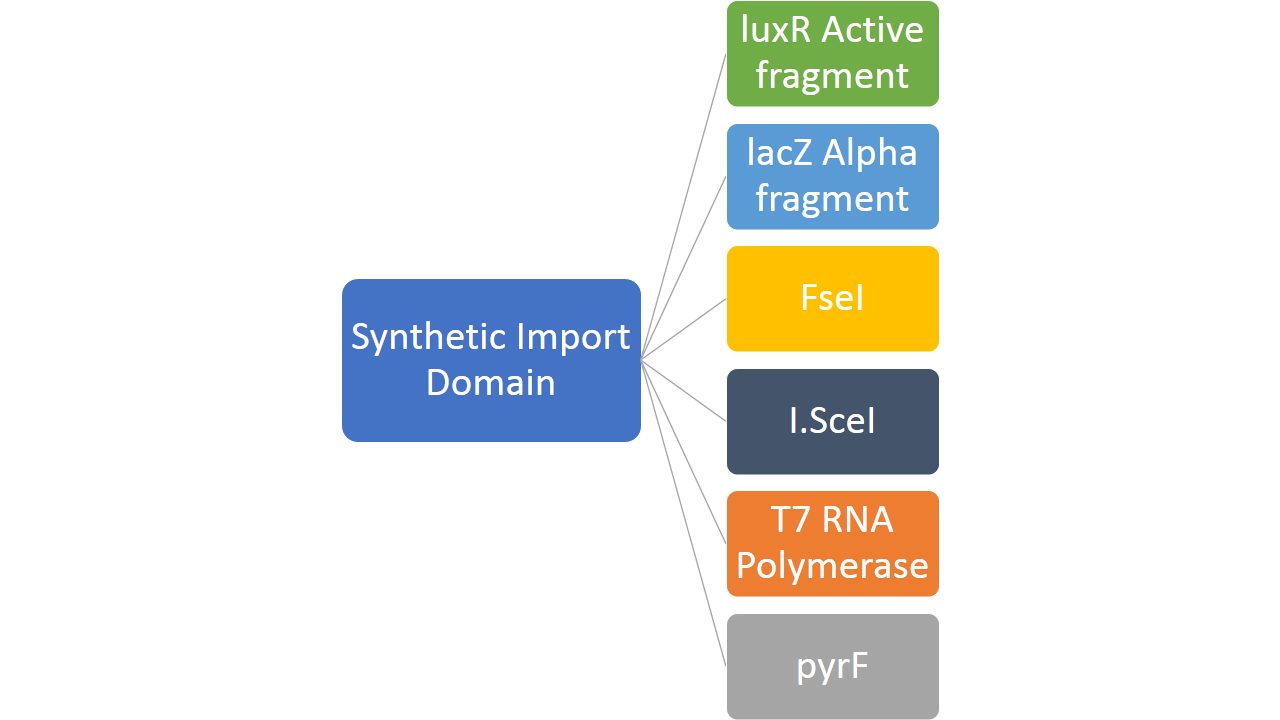
In addition to this, we fused restriction enzymes FseI and I-SceI both to the colicin E2 and colicin D "synthetic import domain". These two enzymes are used in different parts of our project like Restriction Enzyme and MAGE part. We also fused a couple of other proteins to easily test the plausibility of our "synthetic import domain" system. So far we managed to build the majority of our constructs but the experimental confirmation is still lacking.
Objectives
- Creating two types of "synthetic import domains" based on colicin E2 and colicin D binding and translocation mechanisms
- Creating new colicin-like toxin by fusing colicin E2 based "synthetic import domain" with RNAse domain of colicin D
- Creating new import mechanism of proteins into bacterial cell by fusing proteins like FseI, I-SceI, LuxR active fragment, LacZ alpha fragment, PyrF and T7 RNA polymerase with the two types of "synthetic import domains"
- Testing and characterising such a novel mechanism of import
Design
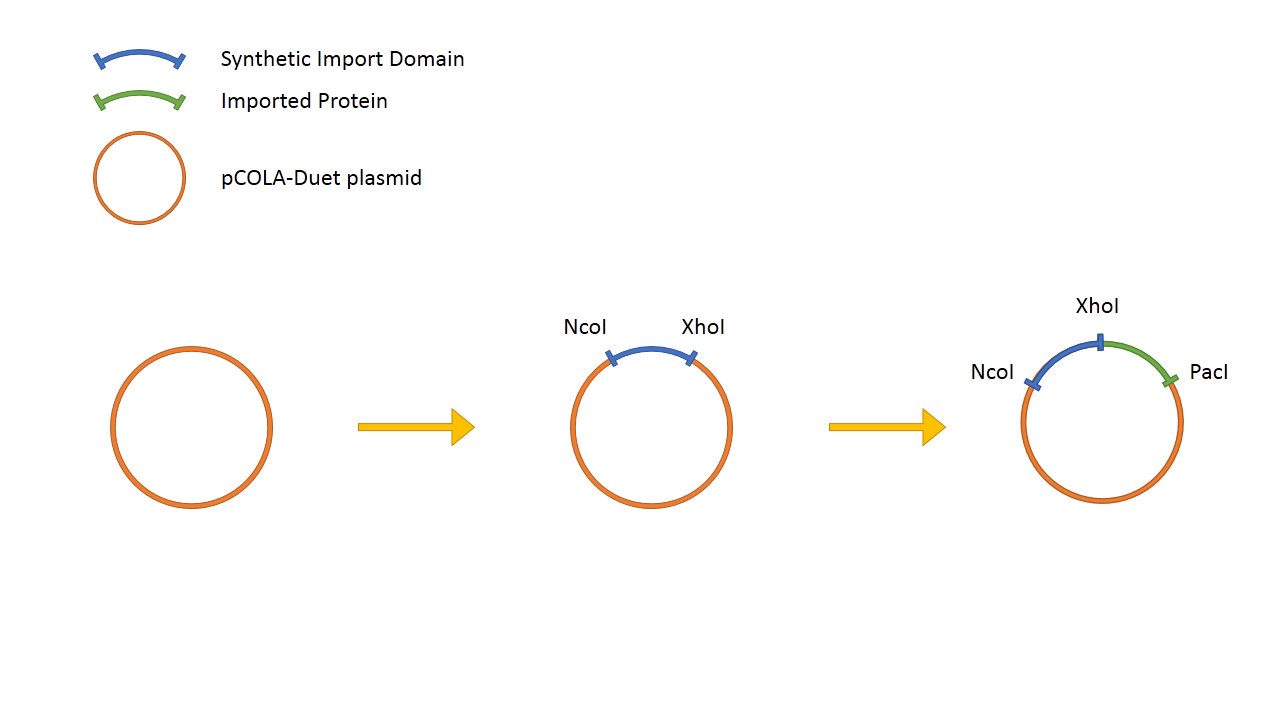
The general design of the Synthetic Import Domain system is very simple. We take the binding and translocation domain from the wild-type colicin and clone it on a plasmid as "Synthetic Import Domain". After, we can fuse any protein of our choice and test if it is going to be imported into other E. coli cells.
Experiments and results
We did not have a chance to test our novel system. So far we fused a majority of our proteins and obtained all the necessary strains for testing our system and doing relevant controls. At this point testing and characterisation of our system is ready to go.
References
- Cascales E, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71:158–229.
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment