Team:HokkaidoU Japan/Notebook/aggregation Week 9
From 2012.igem.org
(28th) |
|||
| (49 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 27th== | + | ===August 27th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| + | ====Aggregation check==== | ||
| + | We found a lot of colonies on LBA plate, spreaded at August 26th. | ||
| + | We incubated 3 colonies in LBAK solution including 1% L-arabinose for 18 hrs. | ||
| + | However, we could not find expression of Ag43 protein. | ||
| - | ==Colony PCR== | + | ====Plasmid extraction==== |
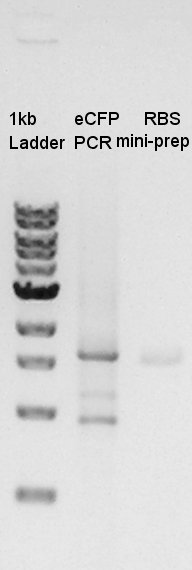
| - | + | We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions. | |
| + | [[image:HokkaidoU2012_120827_take-iroiro.jpg|thumb|Plasmid extraction resulsts]] | ||
| + | |||
| + | ====Colony PCR==== | ||
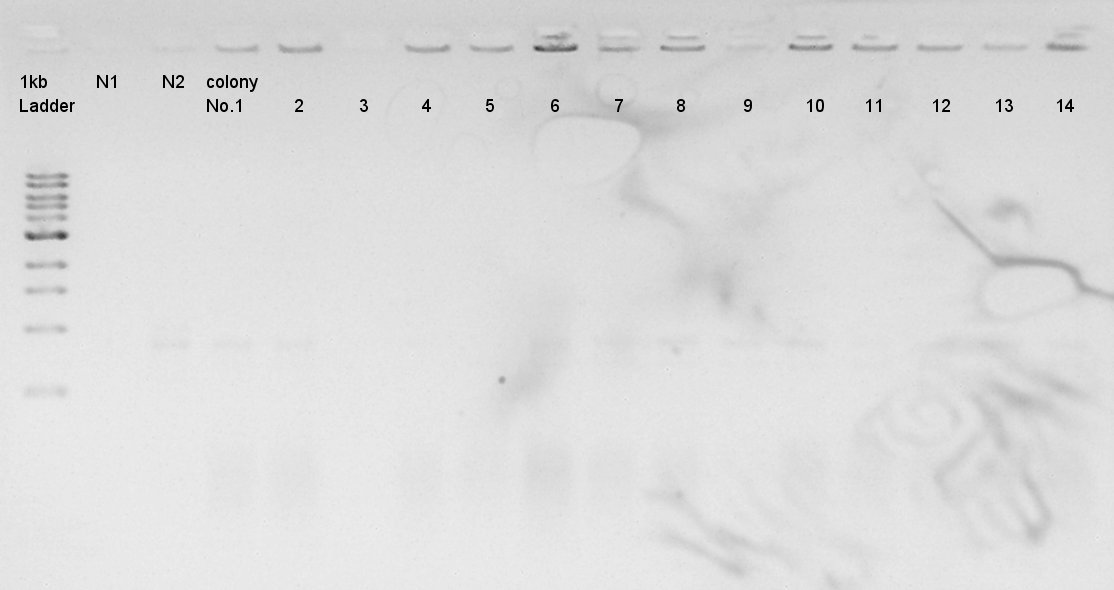
Colony PCR to confirm that whether the pT7-RBS-Ag43-dT on pSB1C3 was successfully ligated or not. | Colony PCR to confirm that whether the pT7-RBS-Ag43-dT on pSB1C3 was successfully ligated or not. | ||
| - | |||
| - | |||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 66: | Line 71: | ||
Desired product is about 695bp. | Desired product is about 695bp. | ||
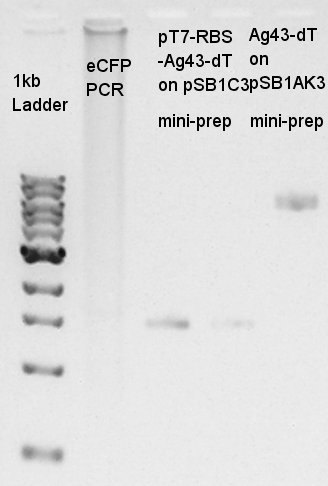
| - | [[image:|thumb|Colony PCR result]] | + | [[image:HokkaidoU2012 120827 colop pT7-RBS-Ag43-dT on pSB1C3.jpg|thumb|Colony PCR result]] |
| - | + | The result did not show the band clearly. We selected No.1 and 2 colony for incubation. | |
| - | + | ====Incubation for plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3==== | |
| - | ==Incubation for | + | |
| - | + | ||
Incubation of pT7-RBS-Ag43-dT on pSB1C3 in LBC liquid medium. | Incubation of pT7-RBS-Ag43-dT on pSB1C3 in LBC liquid medium. | ||
#Prepared 2 ml LBC into culture tubes. | #Prepared 2 ml LBC into culture tubes. | ||
#Re-suspended 2 colonies (No.1 and No.2 respectively). | #Re-suspended 2 colonies (No.1 and No.2 respectively). | ||
#Incubated at 37C for 15 hrs. | #Incubated at 37C for 15 hrs. | ||
| - | |||
| - | ==Transformation of ptet-RBS(B0034)-CFP-dT on pSB1A2== | + | ====Transformation of ptet-RBS(B0034)-CFP-dT on pSB1A2==== |
| - | + | ||
#Mixed 1 ul DNA to 50 ul of thawed competent cells on ice. | #Mixed 1 ul DNA to 50 ul of thawed competent cells on ice. | ||
#Incubated on ice for 30 min. | #Incubated on ice for 30 min. | ||
| Line 89: | Line 90: | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
#Incubated the plates at 37C for 15 hours. | #Incubated the plates at 37C for 15 hours. | ||
| - | |||
| - | ==PCR of RBS-YFP-dT== | + | ====PCR of RBS-YFP-dT==== |
| - | + | Amplified the construct with 100bp-up-EX primer and 200bp-down-PS primer. | |
| - | Amplified the construct | + | |
Mixed PCR solutions. | Mixed PCR solutions. | ||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|Solution | |Solution | ||
| - | |Volume(ul) | + | |Volume (ul) |
|- | |- | ||
|DNA | |DNA | ||
| Line 155: | Line 154: | ||
Cycle:2~4 x 35 | Cycle:2~4 x 35 | ||
| - | </ | + | <br style="line-height: 0; clear: both;" /> |
</div></div> | </div></div> | ||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===August 28th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | |||
| + | ====Digestion==== | ||
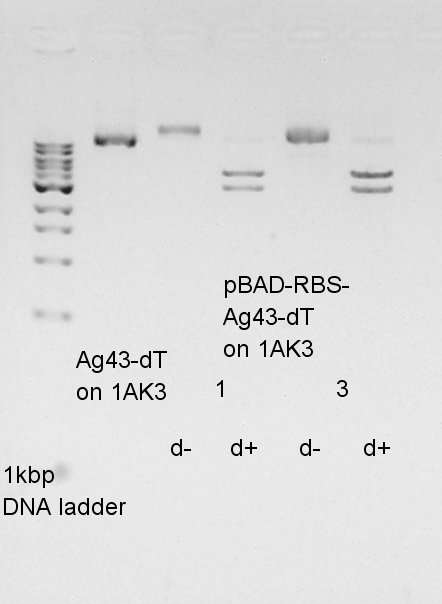
| + | We digested plasmid extraction products (pBAD-RBS-Ag43-dT on pSB1AK3) by EcoRI and SpeI. | ||
| + | Because I'd like to check the size of insert and vector of getting plasmid. | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution | ||
| + | |5 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |11 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |180 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
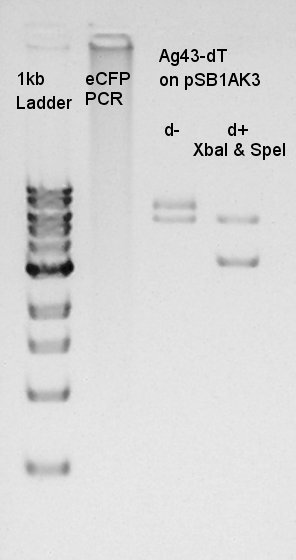
| + | [[image:HokkaidoU2012_120828_pbad-rbs-ag43-dt-dig.jpg|thumb|digestion result]] | ||
| + | |||
| + | ====Gel extraction==== | ||
| + | Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | ||
| + | |||
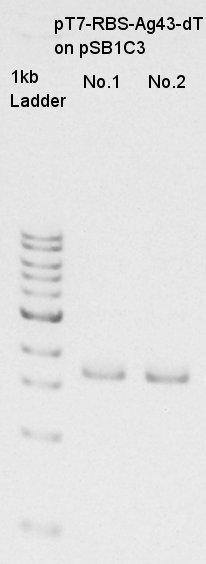
| + | ====Plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3==== | ||
| + | Plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3. We re-suspended No.1,2 colonies and incubated. | ||
| + | |||
| + | [[image:HokkaidoU2012 120828 mini-prep pT7-RBS-Ag43-dT No.jpg|thumb|Plasmid extraction result]] | ||
| + | |||
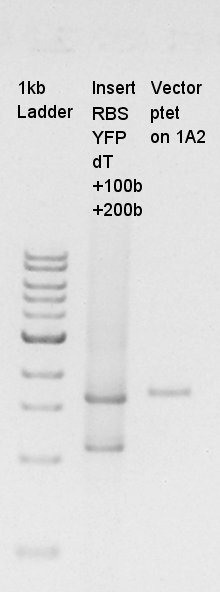
| + | ====Estimation of Concentration of RBS-eYFP-dT and ptetR-pSB1A2==== | ||
| + | {| | ||
| + | |Solution | ||
| + | |Value (ul) | ||
| + | |- | ||
| + | |1kb ladder | ||
| + | |1 | ||
| + | |- | ||
| + | |Insert | ||
| + | |1 | ||
| + | |- | ||
| + | |Vector | ||
| + | |1 | ||
| + | |- | ||
| + | |} | ||
| + | [[image:HokkaidoU2012 120828 Vector-ptetonpSB1C3-Concentrationcheck Insert-rbs-yfp-dt-PCR.jpg|thumb|Electrophoresis result]] | ||
| + | |||
| + | From this result, We estimated that the concentration of Insert DNA solution is about 40 ng/ul, and Vector DNA is also about 40 ng/ul. | ||
| + | |||
| + | ====Digestion of ptet-pSB1A2 and RBS-Ag43-dT==== | ||
| + | ptet-pSB1A2(Vector) was digested with SpeI and PstI, and RBS-Ag43-dT(Insert) was cut with XbaI and PstI.<br /> | ||
| + | Vector | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (40 ng/ul) | ||
| + | |5 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |PstI | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |12 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Inset | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (40 ng/ul) | ||
| + | |14 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |PstI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |2 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |180 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[image:HokkaidoU2012 120828 Digestion Insert-rbs-yfp-dt-PCR(XP) Vector-ptetonpSB1C3(SP).jpg|thumb|digestion result]] | ||
| + | From this result, we confirmed that Insert and Vector DNA were digested. | ||
| + | |||
| + | ====Ethanol Precipitation==== | ||
| + | Ethanol precipitation for ptet-pSB1A2 and RBS-eYFP-dT digestion products. | ||
| + | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 15 min at 4C. | ||
| + | #Removed supernatant and added 220 ul of 70% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 10 min at 4C. | ||
| + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| + | |||
| + | ====Ligation==== | ||
| + | Ligation for ptet-pSB1A2 and RBS-eYFP-dT. | ||
| + | We used Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer. | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Vector DNA | ||
| + | |3 ul | ||
| + | |- | ||
| + | |Insert DNA | ||
| + | |3 ul | ||
| + | |- | ||
| + | |Ligation Mighty Mix | ||
| + | |6 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |12 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Ligation reaction time was in detail below. | ||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |16 | ||
| + | |30 | ||
| + | |- | ||
| + | |65 | ||
| + | |10 | ||
| + | |- | ||
| + | |4 | ||
| + | |Hold | ||
| + | |} | ||
| + | |||
| + | ====Transformation==== | ||
| + | Transformation for ligation product (ptet-RBs-eYFP-dT on pSB1A2) into DH5α. | ||
| + | #Mixed 1 ul of DNA to 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Mixed 350 ul of LB. | ||
| + | #Prepared and Labeled two plastic plates with LBA. | ||
| + | #Plated 300 ul of the culture onto first dish and spread. | ||
| + | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| + | #Incubated the plates at 37C for 15 hrs. | ||
| + | |||
| + | ====Gel extraction product check of pT7-RBS-Ag43-dT on pSB1C3==== | ||
| + | We couldn't get desired plasmid DNA by transformation. We doubted contamination of non-digested vector DNA and decided to test the gel extraction product of pT7-RBS on pSB1C3, which were successfully separated from non-digested product or not by transformation. | ||
| + | #Mixed 1 ul of each DNA of ligation product and digestion product to 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Mixed 350 ul of LB. | ||
| + | #Incubated for 2 hrs to get the resistance to Chloramphenicol. | ||
| + | #Prepared and Labeled two plastic plates with LBC. | ||
| + | #Plated 300 ul of the culture onto first dish and spread. | ||
| + | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| + | #Incubated the plates at 37C for 16 hrs. | ||
| + | |||
| + | There were no colony on the LBC plate that was spread solution mixed digestion product. | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | </div></div> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==August 28th== | + | ===August 29th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==mini-prep of pT7-RBS-Ag43-dT on pSB1C3== | + | ====Aggregation check==== |
| - | + | We incubated 9 colonies transformed at August 25th in LBAK solution including 1% L-arabinose for 16 hrs. | |
| - | < | + | Then we find that 1 culture expressing Ag43. |
| + | Observed the E. coli cluster by a microscope of 100 magnifications. | ||
| + | |||
| + | |||
| + | Then we checked whether pBAD promoter is behaving exactly or not. | ||
| + | #1 colony ( could expressing Ag43 ) incubated in 2 ml LB at 37C for 2 hrs. | ||
| + | #Prepared 5 kinds of LB medium differing the concentration of L-arabinose. | ||
| + | <br />No. 1 :0.001% | ||
| + | <br />No. 2 :0.1% | ||
| + | <br />No. 3 :0.4% | ||
| + | <br />No. 4 :1.0% | ||
| + | <br />No. 5 :0%<br /> | ||
| + | #Added 400 ul of culture in several LB medium. | ||
| + | #Incubated for 17 hrs and 30 min at 37C. | ||
| + | |||
| + | ====PCR of eCFP(E0020)==== | ||
| + | Amplified the part with 100bp-up-EX primer and 200bp-down-PS primer. | ||
| + | Desired product is about 800~900bp.,br />,br /> | ||
| + | Mixed PCR solutions. | ||
| + | {|class="hokkaidou-table-pcr-reagent" | ||
| + | |Solution | ||
| + | |Volume (ul) | ||
| + | |- | ||
| + | |DNA | ||
| + | |1 | ||
| + | |- | ||
| + | |100bp-up-EX | ||
| + | |1 | ||
| + | |- | ||
| + | |200bp-down-PS | ||
| + | |1 | ||
| + | |- | ||
| + | |MgSO4 | ||
| + | |3 | ||
| + | |- | ||
| + | |dNTP | ||
| + | |5 | ||
| + | |- | ||
| + | |10x KOD-Plus-Neo Buffer | ||
| + | |5 | ||
| + | |- | ||
| + | |KOD-Plus-Neo | ||
| + | |1 | ||
| + | |- | ||
| + | |DW | ||
| + | |33 | ||
| + | |- | ||
| + | |Total | ||
| + | |50 | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-time" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Second | ||
| + | |- | ||
| + | |1 | ||
| + | |94 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |98 | ||
| + | |10 | ||
| + | |- | ||
| + | |3 | ||
| + | |58.2 | ||
| + | |30 | ||
| + | |- | ||
| + | |4 | ||
| + | |68 | ||
| + | |60 | ||
| + | |- | ||
| + | |5 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | Cycle:2~4 x 35 | ||
| + | |||
| + | ====Colony PCR==== | ||
| + | Colony PCR of pT7-RBS-Ag43-dT on pSB1C3 transformed at August 28th. | ||
| + | |||
| + | {|class="hokkaidou-table-pcr-reagent" | ||
| + | |- | ||
| + | |DNA solution | ||
| + | |4 ul | ||
| + | |- | ||
| + | |Kapa-Taq(Taq polymerase) | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Forward Primer(ag43-f4 primer) | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |Reverse Primer(200bp down primer) | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |10 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-time" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Second | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |30 | ||
| + | |- | ||
| + | |3 | ||
| + | |53.0 | ||
| + | |30 | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |6 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | Cycle:2~4 x 35 | ||
| + | |||
| + | We used N1 (DW only) and N2 (Ag43-dT on pSB1AK3) as controls. | ||
| + | Desired product is about 695bp. This length is almost same as N2. | ||
| + | |||
| + | [[image:HokkaidoU2012 120829 colop pT7-RBs-Ag43-dTonpSB1C3.jpg|thumb|Colony PCR result]] | ||
| + | |||
| + | We decided to incubate the colony of No.7 and 8. | ||
| + | |||
| + | ====Incubation for plasmid extraction of pT7-RBS-Ag43-dTonpSB1C3 and Ag43-dTonpSB1AK3==== | ||
| + | Incubation of pT7-RBS-Ag43-dTonpSB1C3 (and Ag43-dT on pSB1AK3) in LBC (LBA) liquid medium. | ||
| + | #Prepared 2 ml LBC (LBA) into culture tubes. | ||
| + | #Re-suspended 2 colonies No.7 and No.8 respectively. Ag43-dT on pSB1AK3 was re-suspended the N2 colony. | ||
| + | #Incubated at 37C for 17 hrs. | ||
| + | |||
| + | ====Estimation of Concentration of RBS-eYFP-dT (PCR product) and ptetR-pSB1A2==== | ||
| + | {| | ||
| + | |Solution | ||
| + | |Value (ul) | ||
| + | |- | ||
| + | |1kb ladder | ||
| + | |1 | ||
| + | |- | ||
| + | |Insert | ||
| + | |1 | ||
| + | |- | ||
| + | |Vector | ||
| + | |1 | ||
| + | |- | ||
| + | |} | ||
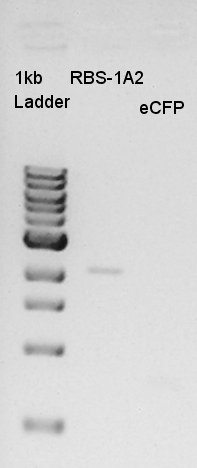
| + | [[image:HokkaidoU2012 120829 PCR 100bpup-eCFP-200bpdown mini-prep B0034.jpg|thumb|Electrophoresis result]] | ||
| + | From this result, We estimated that the concentration of Insert DNA solution is about 36 ng/ul, and Vector DNA is also about 29 ng/ul. | ||
| + | |||
| + | ====Digestion of ptet-pSB1A2 and RBS-Ag43-dT==== | ||
| + | ptet-pSB1A2(Vector) was digested with EcoRI and SpeI, and RBS-Ag43-dT(Insert) was cut with EcoRI and XbaI. | ||
| + | And we used construct of pT7-RBS on pSB1C3 as control for the function of XbaI.<br /> | ||
| + | Insert (eCFP) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (36 ng/ul) | ||
| + | |5 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | Vector(RBS on pSB1A2) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (29 ng/ul) | ||
| + | |6 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |10 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | control (pT7-RBs on pSB1C3) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (30 ng/ul) | ||
| + | |6 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |100x BSA | ||
| + | |0.2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |11 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[image:HokkaidoU2012 120829 Digestion eCFP RBSonpSB1A2 XbaIcontrol.jpg|thumb|digestion result]] | ||
| + | |||
| + | From this result, vector DNA were digested by XbaI, but remains some amount of non-digested DNA. | ||
| + | And insert DNA solution were not existed in both d- and d+ lines.It means we failed to extract from gel after electrophoresis. We decided to retry PCR. | ||
| + | |||
| + | ====PCR of eCFP(E0020)==== | ||
| + | Amplified the part with 100bp-up-EX primer and 200bp-down-PS primer. | ||
| + | Desired product is about 800~900bp.<br /><br /> | ||
| + | Mixed PCR solutions. | ||
| + | {|class="hokkaidou-table-pcr-reagent" | ||
| + | |Solution | ||
| + | |Volume (ul) | ||
| + | |- | ||
| + | |DNA | ||
| + | |1 | ||
| + | |- | ||
| + | |100bp-up-EX | ||
| + | |1 | ||
| + | |- | ||
| + | |200bp-down-PS | ||
| + | |1 | ||
| + | |- | ||
| + | |MgSO4 | ||
| + | |3 | ||
| + | |- | ||
| + | |dNTP | ||
| + | |5 | ||
| + | |- | ||
| + | |10x KOD-Plus-Neo Buffer | ||
| + | |5 | ||
| + | |- | ||
| + | |KOD-Plus-Neo | ||
| + | |1 | ||
| + | |- | ||
| + | |DW | ||
| + | |33 | ||
| + | |- | ||
| + | |Total | ||
| + | |50 | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-time" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Second | ||
| + | |- | ||
| + | |1 | ||
| + | |94 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |98 | ||
| + | |10 | ||
| + | |- | ||
| + | |3 | ||
| + | |58.0 | ||
| + | |30 | ||
| + | |- | ||
| + | |4 | ||
| + | |68 | ||
| + | |60 | ||
| + | |- | ||
| + | |5 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | Cycle:2~4 x 35 | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===August 30th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Estimation of concentration of eCFP, pT7-RBS-Ag43-dT-pSB1C3 and Ag43-dT-pSB1AK3==== | ||
| + | No. 7,8 means the colony number of colony PCR of pT7-RBS-Ag43-dT-pSB1C3. | ||
| + | |||
| + | [[image:HokkaidoU2012 120830 PCR-eCFP mini-prep pT7-RBS-Ag43-dTonpSB1C3 No.jpg|thumb|electrophoresis result]] | ||
| + | |||
| + | We failed the PCR for E0020. | ||
| + | We estimated the concentration and the result was that pT7-RBS-Ag43-dT-pSB1C3 is 31 ng/ul and Ag43-dT-pSB1AK3 is 35 ng/ul. | ||
| + | |||
| + | ====Digestion of pT7-RBS-pSB1C3 and Ag43-dT-pSB1AK3==== | ||
| + | Digestion of pT7-RBS-pSB1C3 (as Vector) and Ag43-dT-pSB1AK3 (as Insert) to make pT7-RBs-Ag43-dT-pSB1C3 construct. | ||
| + | <br /> | ||
| + | Vector | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (31 ng/ul) | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xm buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |16 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Inset | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (35 ng/ul) | ||
| + | |14 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |2 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | As a background, we did PCR for eCFP again. | ||
| + | [[image:HokkaidoU2012 120830 PCR-eCFP digestion-Ag43-dTonpSB1AK3.jpg|thumb|digestion and PCR result]] | ||
| + | |||
| + | We did not do digestion of vector DNA sokution, result, because it has only too low concentration to be digested product solution. | ||
| + | <br /> | ||
| + | We failed PCR again. | ||
| + | The digestion result was confirmed that the Insert DNA was successfully digested by XbaI and SpeI. | ||
| + | And we extracted the insert DNA digestion product from agarose gel by gel-extraction kit and got 20 ul of Insert DNA solution. | ||
| + | |||
| + | ====Digestion of pSB1AK3 by hindIII==== | ||
| + | The gel-extracted DNA solution contains Ag43-dT DNA and pSB1AK3 DNA because these DNA have same size of base pair and exist as same band in agarose gel. Thus we digest pSB1AK3 with HindIII to separate it from Ag43-dT. | ||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (35 ng/ul) | ||
| + | |20 ul | ||
| + | |- | ||
| + | |HindIII | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |3 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |7 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |180 | ||
| + | |- | ||
| + | |2 | ||
| + | |70 | ||
| + | |10 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | As a background, we did PCR for eCFP again. | ||
| + | |||
| + | |||
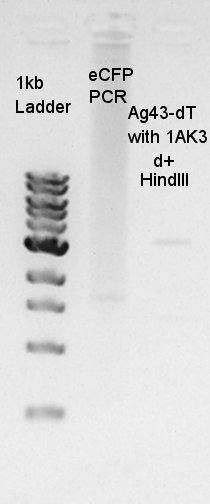
| + | [[image:HokkaidoU2012 120830 PCR-eCFP digestion(HindIII)-Ag43-dTonpSB1AK3.jpg|thumb|PCR and digestion result]] | ||
| + | |||
| + | We failed the PCR for E0020. | ||
| + | The digestion result was confirmed that the insert DNA was successfully digested with HindIII. | ||
| + | |||
| + | ====Ethanol precipitation of pT7-RBS-Ag43-dT-pSB1C3 and Ag43-dT==== | ||
| + | We did ethanol precipitation to get more high concentration DNA solution of each part. And we got 5ul of each DNA solution. | ||
| + | [[image:HokkaidoU2012 120831 Insert-d--d+ Vector-d+.jpg|thumb|ethanol precipitation result]] | ||
| + | The electrophoresis result showed that the pT7-RBS-pSB1C3 construct was successfully digested by SpeI and get enough concentration of DNA solution to ligate. | ||
| + | |||
| + | ====Ligation of pT7-RBS-pSB1C3 and Ag43-dT==== | ||
| + | Ligation of pT7-RBS-pSB1C3 (as Vector) and Ag43-dT (as Insert). | ||
| + | We used Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer. | ||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Vector DNA | ||
| + | |2.5 ul | ||
| + | |- | ||
| + | |Insert DNA | ||
| + | |2.5 ul | ||
| + | |- | ||
| + | |Ligation Mighty Mix | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |10 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Ligation reaction time was in detail below. | ||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |16 | ||
| + | |30 | ||
| + | |- | ||
| + | |65 | ||
| + | |10 | ||
| + | |- | ||
| + | |4 | ||
| + | |Hold | ||
| + | |} | ||
| + | |||
| + | [[image:HokkaidoU2012 120831 Ligation Vector Insert Ligationproduct(4ul).jpg|thumb|ligation result]] | ||
| + | |||
| + | ====Incubation for plasmid extraction of Ag43-dT-pSB1AK3 and ptet-RBS-YFP-dT-pSB1A2==== | ||
| + | Incubation of Ag43-dT-pSB1AK3 and ptet-RBS-YFP-dT-pSB1A2 in LBA liquid medium. | ||
| + | #Prepared 2 ml LBC into culture tubes. | ||
| + | #Re-suspended 1 colony from each plate that each construct plated. | ||
| + | #Incubated at 37C for 22 hrs (Ag43-dT-pSB1AK3 was incubated for 41 hrs) | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===August 31st=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Plasmid extraction for pBAD-RBS-Ag43-dT on pSB1AK3==== | ||
| + | We used promega SV miniprep kit. | ||
| + | We got 100 ul DNA solution. | ||
| + | |||
| + | [[image:HokkaidoU2012_120831_pbad-rbs-ag43-dt-on1ak3.jpg|thumb|plasmid extraction result]] | ||
| + | |||
| + | ====Digestion==== | ||
| + | We digested plasmid extraction products (pBAD-RBS-Ag43-dT on pSB1AK3) with EcoRI and SpeI. | ||
| + | Because I'd like to check the size of insert and vector of getting plasmid. | ||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution | ||
| + | |6 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |10 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[image:HokkaidoU2012_120831_take-iroiro2.jpg|thumb|digestion result]] | ||
| + | |||
| + | ====Transformation of pT7-RBS-Ag43-dT-pSB1C3 and eCFP (E0020)==== | ||
| + | #Mixed 2 ul ligation product and 1ul E0020 DNA solution to each 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Mixed 350 ul of LB. | ||
| + | #Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA and LBC). | ||
| + | #Incubated for 2 hrs to get the resistance to Chloramphenicol (E0020 mixture skipped this step). | ||
| + | #Plated 300 ul of the culture onto first dish and spread. | ||
| + | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| + | #Incubated the plates at 37C for 15 hrs. | ||
| + | |||
| + | We failed to transformation of pT7-RBS-Ag43-dT-pSB1C3. | ||
| + | |||
| + | ====Confirmation of amplification of ptet-RBS-eYFP-dT-pSB1A2==== | ||
| + | After plasmid extraction, we confirmed whether the desired construct ptet-RBS-eYFP-dT was really amplified or not by PCR. | ||
| + | We chose EX-F primer as forward primer and PS-R primer as reverse primer then amplified about 1000bp of insert DNA. | ||
| + | As controls, we chose DW as N1 and ptet-RBS-eCFP-dT construct as N2. | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-reagent" | ||
| + | |- | ||
| + | |DNA solution | ||
| + | |4 ul | ||
| + | |- | ||
| + | |Kapa-Taq(Taq polymerase) | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Forward Primer(EX-F primer) | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |Reverse Primer(PS-R primer) | ||
| + | |0.5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |10 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-time" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Second | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |30 | ||
| + | |- | ||
| + | |3 | ||
| + | |68.9 | ||
| + | |30 | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |6 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | Cycle:2~4 x 35 | ||
| + | |||
| + | [[image:HokkaidoU2012 120831 colop N1(DW) N2(ptet-RBS-eCFP-dT) 1(ptet-RBS-eYFP-dT).jpg|thumb|PCR result]] | ||
| + | |||
| + | This image showed that the desired construct (about 1000 bp) amplified by incubation. | ||
| + | |||
| + | ====Incubation for plasmid extraction of eCFP-pSB1A2==== | ||
| + | Incubation of eCFP-pSB1A2 in LBA liquid medium. | ||
| + | #Prepared 2 ml LBA into culture tubes. | ||
| + | #Re-suspended 1 colony. | ||
| + | #Incubated at 37C for 16 hrs. | ||
| + | |||
| + | ====Transformation of ptet-RBS-eYFP-dT-pSB1A2 and pT7-RBS-Ag43-dT-pSB1C3==== | ||
| + | #Mixed 2 ul pT7-RBS-Ag43-dT-pSB1C3 ligation product and 1ul ptet-RBS-eYFP-dT-pSB1A2 DNA solution to each 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Mixed 350 ul of LB. | ||
| + | #Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA and LBC). | ||
| + | #Incubated for 2 hrs to get the resistance to Chloramphenicol (ptet-RBS-eYFP-dT-pSB1A2 mixture skipped this step). | ||
| + | #Plated 300 ul of the culture onto first dish and spread. | ||
| + | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| + | #Incubated the plates at 37C for hrs. | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 1st=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Digestion for pSB1C3==== | ||
| + | We have to change the plasmid backbone to pSB1C3. | ||
| + | We got the insert DNA at August 31th. | ||
| + | So we need to get the pSB1C3 solution digested with EcoRI and SpeI. | ||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |pSB1C3 (60 ng/ul) | ||
| + | |3 ul | ||
| + | |- | ||
| + | |Eco RI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |13 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |180 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | [[image:HokkaidoU2012_120902_take-iroiro.jpg|thumb|digestion result]] | ||
| + | ====Gel extraction==== | ||
| + | Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution. | ||
| + | |||
| + | ====Ethanol precipitation==== | ||
| + | Ethanol precipitation to get more high concentration of pBAD-RBS-Ag43-dT solution and pSB1C3 solution digested by EcoRI & SpeI. | ||
| + | #Added 2 ul of NaoAc, 1.5 ul of glycogen and 50 ul of 100% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 10 min at 4C. | ||
| + | #Removed supernatant and added 220 ul of 70% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 10 min at 4C. | ||
| + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| + | |||
| + | ====Ligation==== | ||
| + | Ligation for insert DNA and pSB1C3. | ||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Vector DNA | ||
| + | |1 ul | ||
| + | |- | ||
| + | |Insert DNA | ||
| + | |4 ul | ||
| + | |- | ||
| + | |Ligation Mighty Mix | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |10 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Ligation reaction time was in detail below. | ||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |16 | ||
| + | |30 | ||
| + | |- | ||
| + | |65 | ||
| + | |10 | ||
| + | |- | ||
| + | |4 | ||
| + | |Hold | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[image:HokkaidoU2012 120825 Ligation pT7-RBS-Ag43-dT on psB1C3.jpg|thumb|Ligation result]] | ||
| + | |||
| + | |||
| + | ====Transformation==== | ||
| + | Transformation for ligation product into DH5α. | ||
| + | #Added 1 ul of DNA to 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Added 350 ul of LB. | ||
| + | #Incubated the cells for 2 hours at 37C. | ||
| + | #Plated 300 ul of the culture onto first LBC dish and spread. | ||
| + | #Added 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second LBC dish and spread. | ||
| + | #Incubated the plates at 37C for 18 hrs. | ||
| + | |||
| + | ====Plasmid extraction of Ag43-dT-pSB1AK3 and RBS-pSB1A2==== | ||
| + | We used plasmid extraction kit of Nippon genetics and got 50 ul Ag43-dT-pSB1AK3 plasmid DNA solution. | ||
| + | |||
| + | ====Digestion of eCFP-pSB1A2 and RBS-pSB1A2==== | ||
| + | To make a construct of eCFP-RBS-pSB1A2, we digested eCFP-pSB1A2 by EcoRI and SpeI, and RBS-pSB1A2 with EcoRI and XbaI. And we digested pT7-RBS-pSB1C3 by XbaI as a control for confirmation of the ability to digest. | ||
| + | <br /> | ||
| + | Insert (eCFP-pSB1A2) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (35 ng/ul) | ||
| + | |11 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |SpeI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xH buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |5 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Vector(RBS-pSB1A2) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (29 ng/ul) | ||
| + | |6 ul | ||
| + | |- | ||
| + | |EcoRI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |10 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | control (pT7-RBS-pSB1C3) | ||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |DNA solution (30 ng/ul) | ||
| + | |6 ul | ||
| + | |- | ||
| + | |XbaI | ||
| + | |1 ul | ||
| + | |- | ||
| + | |10xM buffer | ||
| + | |2 ul | ||
| + | |- | ||
| + | |100x BSA | ||
| + | |0.2 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |11 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-digestion" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |1 | ||
| + | |37 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |60 | ||
| + | |15 | ||
| + | |- | ||
| + | |3 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[image:HokkaidoU2012 120902 Digestion I-+ V-+ C(XbaI)-+.jpg|thumb|digestion result]] | ||
| + | |||
| + | From this image, we confirmed that DNA were digested into fragments and all of restriction enzyme worked. | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | </div></div> | ||
| + | |||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 2nd=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Ethanol precipitation of digestion products (eCFP and RBS-pSB1A2) and estimation of concentration==== | ||
| + | |||
| + | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
| + | #Centrifuged at 14000 rpm, 30 min at 4C. | ||
| + | #Removed supernatant and added 220 ul of 70% ethanol. | ||
| + | #Centrifuged at 15000 rpm, 15 min at 4C. | ||
| + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. | ||
| + | |||
| + | [[image:HokkaidoU2012 120902 Ethapre V I.jpg|thumb|ethanol precipitation result]] | ||
| + | |||
| + | We estimated the concentration of ethanol presipitation products.The concentration of Insert DNA solution is about 20 ng/ul and Vector DNA solution is about 37 ng/ul. | ||
| + | |||
| + | ====Incubation of pT7-RBS-Ag43-dT-pSB1C3 transformation product==== | ||
| + | We failed to cultivation of above construct two times so we tried to incubation in LBC liquid medium. | ||
| + | #Prepared 1.8 ml LBC into culture tubes. | ||
| + | #Re-suspended 200 ul transformation product solution (which was mixed with LB and incubated for 3 hrs). | ||
| + | #Incubated at 37C. | ||
| + | |||
| + | ====Ligation of eCFP and RBS-pSB1A2==== | ||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Vector DNA (37 ng/ul) | ||
| + | |1 ul | ||
| + | |- | ||
| + | |Insert DNA (20 ng/ul) | ||
| + | |4 ul | ||
| + | |- | ||
| + | |Ligation Mighty Mix | ||
| + | |6 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |11 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | Ligation reaction time was in detail below. | ||
| + | |||
| + | {|class="hokkaidou-table-ligation" | ||
| + | |- | ||
| + | |Degree | ||
| + | |Minute | ||
| + | |- | ||
| + | |16 | ||
| + | |30 | ||
| + | |- | ||
| + | |65 | ||
| + | |10 | ||
| + | |- | ||
| + | |4 | ||
| + | |Hold | ||
| + | |} | ||
| + | |||
| + | |||
| + | ====Transformation of eCFP-RBS-pSB1A2==== | ||
| + | #Mixed 2 ul eCFP-RBS-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | ||
| + | #Incubated on ice for 30 min. | ||
| + | #Mixed 350 ul of LB. | ||
| + | #Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA). | ||
| + | #Plated 300 ul of the culture onto first dish and spread. | ||
| + | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| + | #Incubated the plates at 37C for 19 hrs. | ||
| - | + | <br style="line-height: 0; clear: both;" /> | |
| - | |||
</div></div> | </div></div> | ||
Latest revision as of 03:13, 27 September 2012
August 27th
Aggregation check
We found a lot of colonies on LBA plate, spreaded at August 26th. We incubated 3 colonies in LBAK solution including 1% L-arabinose for 18 hrs. However, we could not find expression of Ag43 protein.
Plasmid extraction
We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50 ul of DNA solutions.
Colony PCR
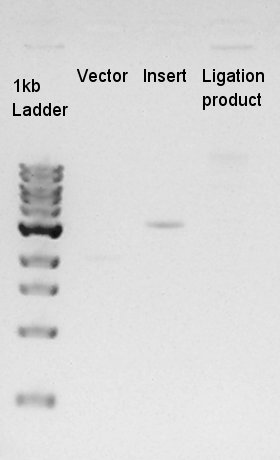
Colony PCR to confirm that whether the pT7-RBS-Ag43-dT on pSB1C3 was successfully ligated or not.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(ag43-f4 primer) | 0.5 ul |
| Reverse Primer(200bp down primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.0 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2 (Ag43-dT on pSB1AK3) as controls. Desired product is about 695bp.
The result did not show the band clearly. We selected No.1 and 2 colony for incubation.
Incubation for plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3
Incubation of pT7-RBS-Ag43-dT on pSB1C3 in LBC liquid medium.
- Prepared 2 ml LBC into culture tubes.
- Re-suspended 2 colonies (No.1 and No.2 respectively).
- Incubated at 37C for 15 hrs.
Transformation of ptet-RBS(B0034)-CFP-dT on pSB1A2
- Mixed 1 ul DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LBC.
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 15 hours.
PCR of RBS-YFP-dT
Amplified the construct with 100bp-up-EX primer and 200bp-down-PS primer. Mixed PCR solutions.
| Solution | Volume (ul) |
| DNA | 1 |
| 100bp-up-EX | 1 |
| 200bp-down-PS | 1 |
| MgSO4 | 3 |
| dNTP | 5 |
| 10x KOD-Plus-Neo Buffer | 5 |
| KOD-Plus-Neo | 1 |
| DW | 33 |
| Total | 50 |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58.2 | 30 |
| 4 | 68 | 60 |
| 5 | 4 | HOLD |
Cycle:2~4 x 35
August 28th
Digestion
We digested plasmid extraction products (pBAD-RBS-Ag43-dT on pSB1AK3) by EcoRI and SpeI. Because I'd like to check the size of insert and vector of getting plasmid.
| DNA solution | 5 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 11 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Gel extraction
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
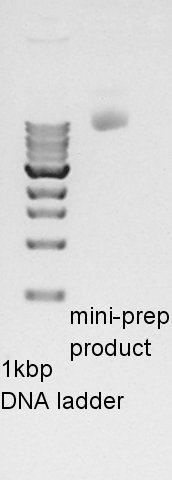
Plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3
Plasmid extraction of pT7-RBS-Ag43-dT on pSB1C3. We re-suspended No.1,2 colonies and incubated.
Estimation of Concentration of RBS-eYFP-dT and ptetR-pSB1A2
| Solution | Value (ul) |
| 1kb ladder | 1 |
| Insert | 1 |
| Vector | 1 |
From this result, We estimated that the concentration of Insert DNA solution is about 40 ng/ul, and Vector DNA is also about 40 ng/ul.
Digestion of ptet-pSB1A2 and RBS-Ag43-dT
ptet-pSB1A2(Vector) was digested with SpeI and PstI, and RBS-Ag43-dT(Insert) was cut with XbaI and PstI.
Vector
| DNA solution (40 ng/ul) | 5 ul |
| SpeI | 0.5 ul |
| PstI | 0.5 ul |
| 10xH buffer | 2 ul |
| DW | 12 ul |
| Total | 20 ul |
Inset
| DNA solution (40 ng/ul) | 14 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 2 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
From this result, we confirmed that Insert and Vector DNA were digested.
Ethanol Precipitation
Ethanol precipitation for ptet-pSB1A2 and RBS-eYFP-dT digestion products.
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
Ligation
Ligation for ptet-pSB1A2 and RBS-eYFP-dT. We used Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer.
| Vector DNA | 3 ul |
| Insert DNA | 3 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 12 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation
Transformation for ligation product (ptet-RBs-eYFP-dT on pSB1A2) into DH5α.
- Mixed 1 ul of DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LBA.
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 15 hrs.
Gel extraction product check of pT7-RBS-Ag43-dT on pSB1C3
We couldn't get desired plasmid DNA by transformation. We doubted contamination of non-digested vector DNA and decided to test the gel extraction product of pT7-RBS on pSB1C3, which were successfully separated from non-digested product or not by transformation.
- Mixed 1 ul of each DNA of ligation product and digestion product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Incubated for 2 hrs to get the resistance to Chloramphenicol.
- Prepared and Labeled two plastic plates with LBC.
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 16 hrs.
There were no colony on the LBC plate that was spread solution mixed digestion product.
August 29th
Aggregation check
We incubated 9 colonies transformed at August 25th in LBAK solution including 1% L-arabinose for 16 hrs. Then we find that 1 culture expressing Ag43. Observed the E. coli cluster by a microscope of 100 magnifications.
Then we checked whether pBAD promoter is behaving exactly or not.
- 1 colony ( could expressing Ag43 ) incubated in 2 ml LB at 37C for 2 hrs.
- Prepared 5 kinds of LB medium differing the concentration of L-arabinose.
No. 1 :0.001%
No. 2 :0.1%
No. 3 :0.4%
No. 4 :1.0%
No. 5 :0%
- Added 400 ul of culture in several LB medium.
- Incubated for 17 hrs and 30 min at 37C.
PCR of eCFP(E0020)
Amplified the part with 100bp-up-EX primer and 200bp-down-PS primer. Desired product is about 800~900bp.,br />,br /> Mixed PCR solutions.
| Solution | Volume (ul) |
| DNA | 1 |
| 100bp-up-EX | 1 |
| 200bp-down-PS | 1 |
| MgSO4 | 3 |
| dNTP | 5 |
| 10x KOD-Plus-Neo Buffer | 5 |
| KOD-Plus-Neo | 1 |
| DW | 33 |
| Total | 50 |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58.2 | 30 |
| 4 | 68 | 60 |
| 5 | 4 | HOLD |
Cycle:2~4 x 35
Colony PCR
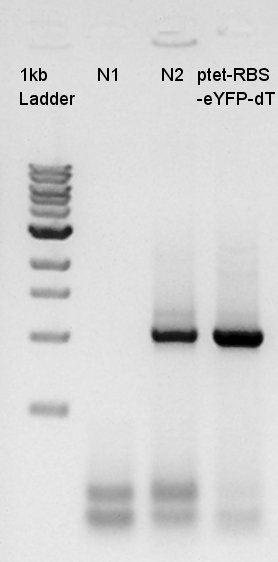
Colony PCR of pT7-RBS-Ag43-dT on pSB1C3 transformed at August 28th.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(ag43-f4 primer) | 0.5 ul |
| Reverse Primer(200bp down primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.0 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2 (Ag43-dT on pSB1AK3) as controls. Desired product is about 695bp. This length is almost same as N2.
We decided to incubate the colony of No.7 and 8.
Incubation for plasmid extraction of pT7-RBS-Ag43-dTonpSB1C3 and Ag43-dTonpSB1AK3
Incubation of pT7-RBS-Ag43-dTonpSB1C3 (and Ag43-dT on pSB1AK3) in LBC (LBA) liquid medium.
- Prepared 2 ml LBC (LBA) into culture tubes.
- Re-suspended 2 colonies No.7 and No.8 respectively. Ag43-dT on pSB1AK3 was re-suspended the N2 colony.
- Incubated at 37C for 17 hrs.
Estimation of Concentration of RBS-eYFP-dT (PCR product) and ptetR-pSB1A2
| Solution | Value (ul) |
| 1kb ladder | 1 |
| Insert | 1 |
| Vector | 1 |
From this result, We estimated that the concentration of Insert DNA solution is about 36 ng/ul, and Vector DNA is also about 29 ng/ul.
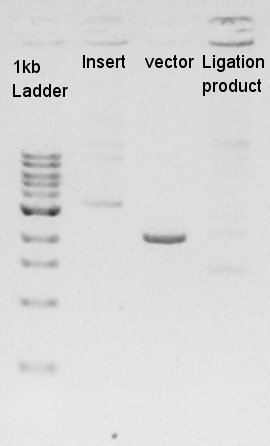
Digestion of ptet-pSB1A2 and RBS-Ag43-dT
ptet-pSB1A2(Vector) was digested with EcoRI and SpeI, and RBS-Ag43-dT(Insert) was cut with EcoRI and XbaI.
And we used construct of pT7-RBS on pSB1C3 as control for the function of XbaI.
Insert (eCFP)
| DNA solution (36 ng/ul) | 5 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 5 ul |
| Total | 20 ul |
Vector(RBS on pSB1A2)
| DNA solution (29 ng/ul) | 6 ul |
| EcoRI | 1 ul |
| XbaI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
control (pT7-RBs on pSB1C3)
| DNA solution (30 ng/ul) | 6 ul |
| XbaI | 1 ul |
| 10xM buffer | 2 ul |
| 100x BSA | 0.2 ul |
| DW | 11 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
From this result, vector DNA were digested by XbaI, but remains some amount of non-digested DNA. And insert DNA solution were not existed in both d- and d+ lines.It means we failed to extract from gel after electrophoresis. We decided to retry PCR.
PCR of eCFP(E0020)
Amplified the part with 100bp-up-EX primer and 200bp-down-PS primer.
Desired product is about 800~900bp.
Mixed PCR solutions.
| Solution | Volume (ul) |
| DNA | 1 |
| 100bp-up-EX | 1 |
| 200bp-down-PS | 1 |
| MgSO4 | 3 |
| dNTP | 5 |
| 10x KOD-Plus-Neo Buffer | 5 |
| KOD-Plus-Neo | 1 |
| DW | 33 |
| Total | 50 |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58.0 | 30 |
| 4 | 68 | 60 |
| 5 | 4 | HOLD |
Cycle:2~4 x 35
August 30th
Estimation of concentration of eCFP, pT7-RBS-Ag43-dT-pSB1C3 and Ag43-dT-pSB1AK3
No. 7,8 means the colony number of colony PCR of pT7-RBS-Ag43-dT-pSB1C3.
We failed the PCR for E0020. We estimated the concentration and the result was that pT7-RBS-Ag43-dT-pSB1C3 is 31 ng/ul and Ag43-dT-pSB1AK3 is 35 ng/ul.
Digestion of pT7-RBS-pSB1C3 and Ag43-dT-pSB1AK3
Digestion of pT7-RBS-pSB1C3 (as Vector) and Ag43-dT-pSB1AK3 (as Insert) to make pT7-RBs-Ag43-dT-pSB1C3 construct.
Vector
| DNA solution (31 ng/ul) | 1 ul |
| SpeI | 1 ul |
| 10xm buffer | 2 ul |
| DW | 16 ul |
| Total | 20 ul |
Inset
| DNA solution (35 ng/ul) | 14 ul |
| XbaI | 1 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 2 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
As a background, we did PCR for eCFP again.
We did not do digestion of vector DNA sokution, result, because it has only too low concentration to be digested product solution.
We failed PCR again.
The digestion result was confirmed that the Insert DNA was successfully digested by XbaI and SpeI.
And we extracted the insert DNA digestion product from agarose gel by gel-extraction kit and got 20 ul of Insert DNA solution.
Digestion of pSB1AK3 by hindIII
The gel-extracted DNA solution contains Ag43-dT DNA and pSB1AK3 DNA because these DNA have same size of base pair and exist as same band in agarose gel. Thus we digest pSB1AK3 with HindIII to separate it from Ag43-dT.
| DNA solution (35 ng/ul) | 20 ul |
| HindIII | 1 ul |
| 10xM buffer | 3 ul |
| DW | 7 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 70 | 10 |
| 3 | 4 | HOLD |
As a background, we did PCR for eCFP again.
We failed the PCR for E0020. The digestion result was confirmed that the insert DNA was successfully digested with HindIII.
Ethanol precipitation of pT7-RBS-Ag43-dT-pSB1C3 and Ag43-dT
We did ethanol precipitation to get more high concentration DNA solution of each part. And we got 5ul of each DNA solution.
The electrophoresis result showed that the pT7-RBS-pSB1C3 construct was successfully digested by SpeI and get enough concentration of DNA solution to ligate.
Ligation of pT7-RBS-pSB1C3 and Ag43-dT
Ligation of pT7-RBS-pSB1C3 (as Vector) and Ag43-dT (as Insert). We used Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer.
| Vector DNA | 2.5 ul |
| Insert DNA | 2.5 ul |
| Ligation Mighty Mix | 5 ul |
| Total | 10 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Incubation for plasmid extraction of Ag43-dT-pSB1AK3 and ptet-RBS-YFP-dT-pSB1A2
Incubation of Ag43-dT-pSB1AK3 and ptet-RBS-YFP-dT-pSB1A2 in LBA liquid medium.
- Prepared 2 ml LBC into culture tubes.
- Re-suspended 1 colony from each plate that each construct plated.
- Incubated at 37C for 22 hrs (Ag43-dT-pSB1AK3 was incubated for 41 hrs)
August 31st
Plasmid extraction for pBAD-RBS-Ag43-dT on pSB1AK3
We used promega SV miniprep kit. We got 100 ul DNA solution.
Digestion
We digested plasmid extraction products (pBAD-RBS-Ag43-dT on pSB1AK3) with EcoRI and SpeI. Because I'd like to check the size of insert and vector of getting plasmid.
| DNA solution | 6 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Transformation of pT7-RBS-Ag43-dT-pSB1C3 and eCFP (E0020)
- Mixed 2 ul ligation product and 1ul E0020 DNA solution to each 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA and LBC).
- Incubated for 2 hrs to get the resistance to Chloramphenicol (E0020 mixture skipped this step).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 15 hrs.
We failed to transformation of pT7-RBS-Ag43-dT-pSB1C3.
Confirmation of amplification of ptet-RBS-eYFP-dT-pSB1A2
After plasmid extraction, we confirmed whether the desired construct ptet-RBS-eYFP-dT was really amplified or not by PCR. We chose EX-F primer as forward primer and PS-R primer as reverse primer then amplified about 1000bp of insert DNA. As controls, we chose DW as N1 and ptet-RBS-eCFP-dT construct as N2.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(EX-F primer) | 0.5 ul |
| Reverse Primer(PS-R primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 68.9 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
This image showed that the desired construct (about 1000 bp) amplified by incubation.
Incubation for plasmid extraction of eCFP-pSB1A2
Incubation of eCFP-pSB1A2 in LBA liquid medium.
- Prepared 2 ml LBA into culture tubes.
- Re-suspended 1 colony.
- Incubated at 37C for 16 hrs.
Transformation of ptet-RBS-eYFP-dT-pSB1A2 and pT7-RBS-Ag43-dT-pSB1C3
- Mixed 2 ul pT7-RBS-Ag43-dT-pSB1C3 ligation product and 1ul ptet-RBS-eYFP-dT-pSB1A2 DNA solution to each 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA and LBC).
- Incubated for 2 hrs to get the resistance to Chloramphenicol (ptet-RBS-eYFP-dT-pSB1A2 mixture skipped this step).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for hrs.
September 1st
Digestion for pSB1C3
We have to change the plasmid backbone to pSB1C3. We got the insert DNA at August 31th. So we need to get the pSB1C3 solution digested with EcoRI and SpeI.
| pSB1C3 (60 ng/ul) | 3 ul |
| Eco RI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 13 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Gel extraction
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
Ethanol precipitation
Ethanol precipitation to get more high concentration of pBAD-RBS-Ag43-dT solution and pSB1C3 solution digested by EcoRI & SpeI.
- Added 2 ul of NaoAc, 1.5 ul of glycogen and 50 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
Ligation
Ligation for insert DNA and pSB1C3.
| Vector DNA | 1 ul |
| Insert DNA | 4 ul |
| Ligation Mighty Mix | 5 ul |
| Total | 10 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation
Transformation for ligation product into DH5α.
- Added 1 ul of DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Added 350 ul of LB.
- Incubated the cells for 2 hours at 37C.
- Plated 300 ul of the culture onto first LBC dish and spread.
- Added 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second LBC dish and spread.
- Incubated the plates at 37C for 18 hrs.
Plasmid extraction of Ag43-dT-pSB1AK3 and RBS-pSB1A2
We used plasmid extraction kit of Nippon genetics and got 50 ul Ag43-dT-pSB1AK3 plasmid DNA solution.
Digestion of eCFP-pSB1A2 and RBS-pSB1A2
To make a construct of eCFP-RBS-pSB1A2, we digested eCFP-pSB1A2 by EcoRI and SpeI, and RBS-pSB1A2 with EcoRI and XbaI. And we digested pT7-RBS-pSB1C3 by XbaI as a control for confirmation of the ability to digest.
Insert (eCFP-pSB1A2)
| DNA solution (35 ng/ul) | 11 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 5 ul |
| Total | 20 ul |
Vector(RBS-pSB1A2)
| DNA solution (29 ng/ul) | 6 ul |
| EcoRI | 1 ul |
| XbaI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
control (pT7-RBS-pSB1C3)
| DNA solution (30 ng/ul) | 6 ul |
| XbaI | 1 ul |
| 10xM buffer | 2 ul |
| 100x BSA | 0.2 ul |
| DW | 11 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
From this image, we confirmed that DNA were digested into fragments and all of restriction enzyme worked.
September 2nd
Ethanol precipitation of digestion products (eCFP and RBS-pSB1A2) and estimation of concentration
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 14000 rpm, 30 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We estimated the concentration of ethanol presipitation products.The concentration of Insert DNA solution is about 20 ng/ul and Vector DNA solution is about 37 ng/ul.
Incubation of pT7-RBS-Ag43-dT-pSB1C3 transformation product
We failed to cultivation of above construct two times so we tried to incubation in LBC liquid medium.
- Prepared 1.8 ml LBC into culture tubes.
- Re-suspended 200 ul transformation product solution (which was mixed with LB and incubated for 3 hrs).
- Incubated at 37C.
Ligation of eCFP and RBS-pSB1A2
| Vector DNA (37 ng/ul) | 1 ul |
| Insert DNA (20 ng/ul) | 4 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 11 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of eCFP-RBS-pSB1A2
- Mixed 2 ul eCFP-RBS-pSB1A2 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 19 hrs.
 "
"