Team:Bielefeld-Germany/Labjournal/week6
From 2012.igem.org
(Difference between revisions)
(→Monday June 4th) |
(→weekly seminar) |
||
| (51 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Bielefeld/Head}} | {{Team:Bielefeld/Head}} | ||
| - | |||
| - | + | <html> | |
| - | + | <style type="text/css"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ul {list-style-image:none;} | ||
| + | #bodyContent{ | ||
| + | background-color: white; | ||
| + | } | ||
| + | </style> | ||
| + | |||
| + | <!-- navigator --> | ||
| + | <div id="nav" class="tabs"> | ||
| + | <div class="scroller"> | ||
| + | <ul style="list-style-type:none"> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/Starting"><strong>Prologue</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week1"><strong>Week 1</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week2"><strong>Week 2</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week3"><strong>Week 3</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week4"><strong>Week 4</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week5"><strong>Week 5</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week6"><strong>Week 6</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week7"><strong>Week 7</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week8"><strong>Week 8</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week9"><strong>Week 9</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week10"><strong>Week 10</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week11"><strong>Week 11</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week12"><strong>Week 12</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week13"><strong>Week 13</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week14"><strong>Week 14</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week15"><strong>Week 15</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week16"><strong>Week 16</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week17"><strong>Week 17</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week18"><strong>Week 18</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week19"><strong>Week 19</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week20"><strong>Week 20</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week21"><strong>Week 21</strong></a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week22"><strong>Week 22</strong></a></li> | ||
| + | |||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <div style="text-align:justify;"> | ||
==Week 6 (06/04 - 06/10/12)== | ==Week 6 (06/04 - 06/10/12)== | ||
| - | === | + | === weekly seminar === |
| + | * As part of the sponsoring with [http://www.merckgroup.com/en/index.html Merck] we will present our project in Darmstadt. Kevin, Nadine and Gabi travel to Darmstadt on 21th of june. | ||
| + | The team will gather on 14th, 15th and 18th of june to practice the presentation. | ||
| + | * Participants at the [http://www.cas.uni-muenchen.de/veranstaltungen/tag_synth_bio_2012/index.html CAS conference for Synthetic Biology]: Hakan, Derya, Miriam, Julia S., Gabi, Malak, Timo, Robert, Isabel, Nils. | ||
| + | * Robert is responsible for ordering the following BioBricks from the Partsregistry: | ||
| + | <partinfo>K500000</partinfo> <br /> | ||
| + | <partinfo>K500001</partinfo> <br /> | ||
| + | <partinfo>K500002</partinfo> <br /> | ||
| + | <partinfo>K500003</partinfo> <br /> | ||
| + | <partinfo>K392014</partinfo> <br /> | ||
| - | * | + | * The treaty with our sponsor [http://www.biocircle.com/en-ca/ BioCircle] is signed. |
| - | * '''Team | + | * All teams presented their lab results. |
| - | ** '''PCR table''' | + | |
| + | ===Monday June 4th=== | ||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | :* The Bacteria ''S. griseus'' and ''S. lavendulae'' has been delivered so we can start with PCRs. We set the first PCR with them as followed: | ||
| + | :** '''PCR table''' | ||
{| class="wikitable" | {| class="wikitable" | ||
|- | |- | ||
| Line 54: | Line 91: | ||
! Temperature !! Time | ! Temperature !! Time | ||
|- | |- | ||
| - | | 1) 98°C || | + | | 1) 98°C || 7 mins |
|- | |- | ||
| - | | 2) 98°C || | + | | 2) 98°C || 20 sec |
|- | |- | ||
| - | | 3) | + | | 3) 55°C || 20 sec |
|- | |- | ||
| 4) 72°C || 1 min | | 4) 72°C || 1 min | ||
| Line 67: | Line 104: | ||
|- | |- | ||
|} | |} | ||
| + | |||
Cycle between step 2 and 4 35 times. | Cycle between step 2 and 4 35 times. | ||
| + | |||
| + | *'''Team Fungal and Plant Laccases: ''' | ||
| + | **Primerdesign for isolating a laccase from ''Arababidopsis thaliana'' cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag. | ||
===Tuesday June 5th=== | ===Tuesday June 5th=== | ||
| - | * '''Team Activity Tests''': After testing the ''T. versicolor'' laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM | + | * '''Team Student Academy:''' |
| + | ** Transformation of a plasmid mixture of either pMTE cp46 His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] or pMTE cp46 His and [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] has a more intense fluorescence and was chosen for the experiment. | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': | ||
| + | ** The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence. | ||
| + | ** Again we did PCRs on ''T. thermophilus'' laccase and ''B. halodurans'' laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction. | ||
| + | |||
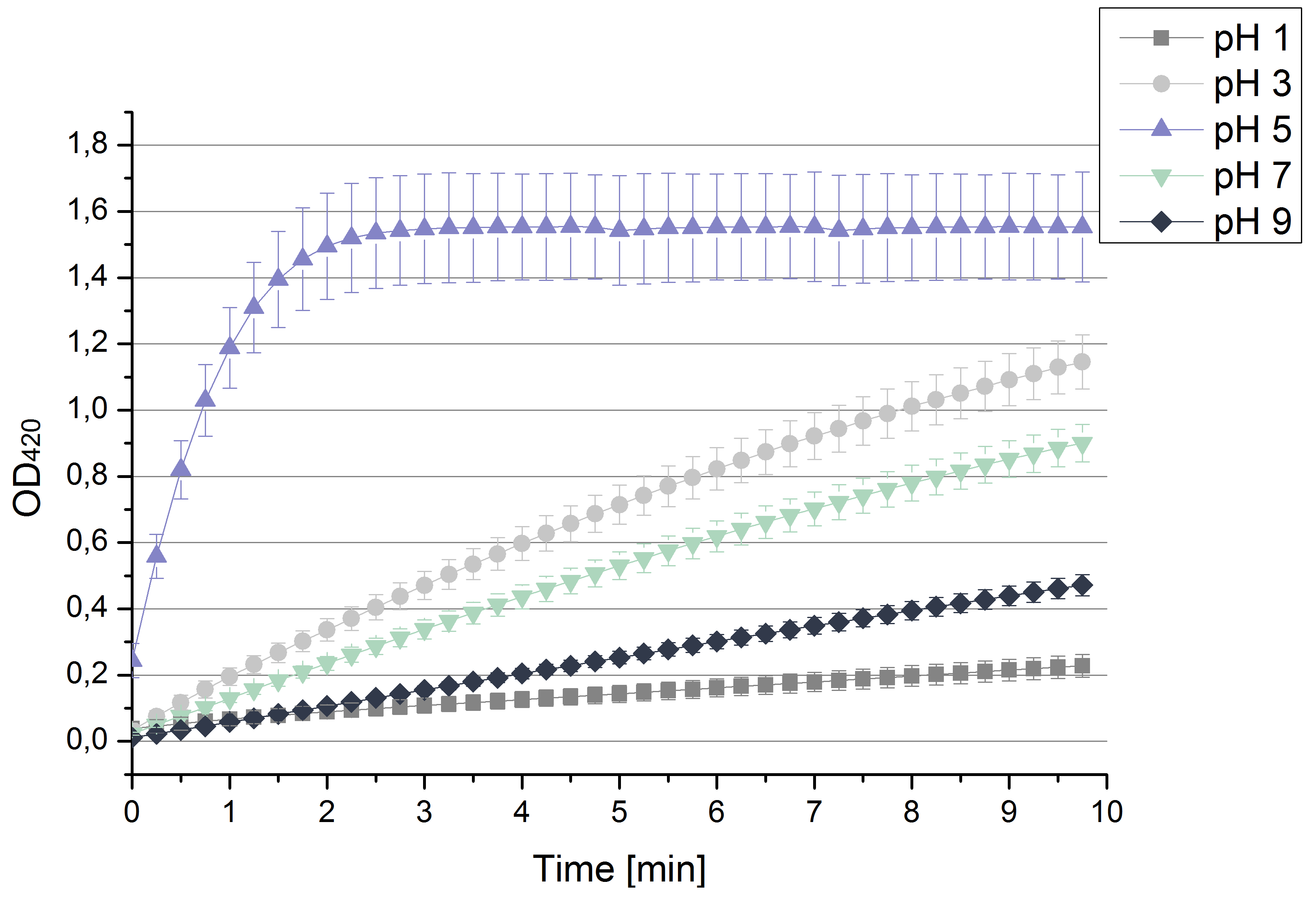
| + | [[File:Bielefeld2012 pHstrametes.png|thumb|right|Activity measurements of the bought laccase from ''T.versiolor'' analyzed through the OD<sub>420</sub> of oxidized ABTS in sodium acetate buffer at different pHs depending on time. Values are calculated by taking the average and standard deviation out of 4 measurements (n=4).]] | ||
| + | * '''Team Activity Tests''': | ||
| + | ** After testing the ''T. versicolor'' laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned. | ||
===Wednesday June 6th=== | ===Wednesday June 6th=== | ||
| Line 77: | Line 127: | ||
** we created all pages and will fill them up with some nice and beautiful content constantly from now on. | ** we created all pages and will fill them up with some nice and beautiful content constantly from now on. | ||
** Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon. | ** Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon. | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases:''' Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent ''E. coli'' KRX cells. | ||
===Thursday June 7th=== | ===Thursday June 7th=== | ||
| - | * '''Team Modeling''': | + | * '''Team Student Academy:''' |
| + | ** Repeating of Transformation of 06/05 to verify the function. It is reproducible :) | ||
| + | |||
| + | * '''Team Cloning of Bacterial Laccases''': Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands. | ||
| + | |||
| + | * '''Team Modeling''': becoming acquainted with matlab while reading the manual | ||
===Friday June 8th=== | ===Friday June 8th=== | ||
| + | |||
| + | *'''Team Cloning of Bacterial Laccases:''' | ||
| + | **We plated colonies for plasmid isolations on new plates and made a control restriction with ''Not''I. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His. | ||
| + | |||
===Saturday June 9th=== | ===Saturday June 9th=== | ||
===Sunday June 10th=== | ===Sunday June 10th=== | ||
| + | |||
| + | |||
| + | {{Team:Bielefeld/Sponsoren}} | ||
Latest revision as of 09:46, 25 September 2012
Contents |
Week 6 (06/04 - 06/10/12)
weekly seminar
- As part of the sponsoring with [http://www.merckgroup.com/en/index.html Merck] we will present our project in Darmstadt. Kevin, Nadine and Gabi travel to Darmstadt on 21th of june.
The team will gather on 14th, 15th and 18th of june to practice the presentation.
- Participants at the [http://www.cas.uni-muenchen.de/veranstaltungen/tag_synth_bio_2012/index.html CAS conference for Synthetic Biology]: Hakan, Derya, Miriam, Julia S., Gabi, Malak, Timo, Robert, Isabel, Nils.
- Robert is responsible for ordering the following BioBricks from the Partsregistry:
<partinfo>K500000</partinfo>
<partinfo>K500001</partinfo>
<partinfo>K500002</partinfo>
<partinfo>K500003</partinfo>
<partinfo>K392014</partinfo>
- The treaty with our sponsor [http://www.biocircle.com/en-ca/ BioCircle] is signed.
- All teams presented their lab results.
Monday June 4th
- Team Cloning of Bacterial Laccases:
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
- PCR table
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
| Material | Volume |
|---|---|
| Buffer (10x Phusion) | 10µL |
| Phusion Polymerase | 0,5µL |
| dNTPs | 1µL |
| Primer Mix | 1µL |
| Template DNA | 1µL |
| DMSO | 1,5µL |
| Water | 35µL |
- PCR program
| Temperature | Time |
|---|---|
| 1) 98°C | 7 mins |
| 2) 98°C | 20 sec |
| 3) 55°C | 20 sec |
| 4) 72°C | 1 min |
| 5) 72°C | 3 min |
| 6) 12°C |
Cycle between step 2 and 4 35 times.
- Team Fungal and Plant Laccases:
- Primerdesign for isolating a laccase from Arababidopsis thaliana cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag.
Tuesday June 5th
- Team Student Academy:
- Transformation of a plasmid mixture of either pMTE cp46 His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] or pMTE cp46 His and [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] has a more intense fluorescence and was chosen for the experiment.
- Team Cloning of Bacterial Laccases:
- The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence.
- Again we did PCRs on T. thermophilus laccase and B. halodurans laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction.
- Team Activity Tests:
- After testing the T. versicolor laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned.
Wednesday June 6th
- Team Wiki: Yay for Team Wiki´s first entry. Our first steps with the iGEM Bielefeld 2012 Wiki contain thinking about contents, layouts, programming and responsibilities. Our first rules are:
- we are programming static pages in HTML and all the other pages (those that will be updated by all team members) in wiki code.
- we created all pages and will fill them up with some nice and beautiful content constantly from now on.
- Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon.
- Team Cloning of Bacterial Laccases: Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent E. coli KRX cells.
Thursday June 7th
- Team Student Academy:
- Repeating of Transformation of 06/05 to verify the function. It is reproducible :)
- Team Cloning of Bacterial Laccases: Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands.
- Team Modeling: becoming acquainted with matlab while reading the manual
Friday June 8th
- Team Cloning of Bacterial Laccases:
- We plated colonies for plasmid isolations on new plates and made a control restriction with NotI. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His.
Saturday June 9th
Sunday June 10th
| 55px | | | | | | | | | | |
 "
"