Team:TU-Delft/Modeling
From 2012.igem.org
| (One intermediate revision not shown) | |
Latest revision as of 03:42, 27 October 2012


Contents |
Overview
We decided to use the modeling expertise of the team members to achieve three key objectives for our project.
- Develop scientific understanding of the yeast pheromone response pathway.
- Test the effects of the changes to the system.
- Aid decision making in the laboratory.
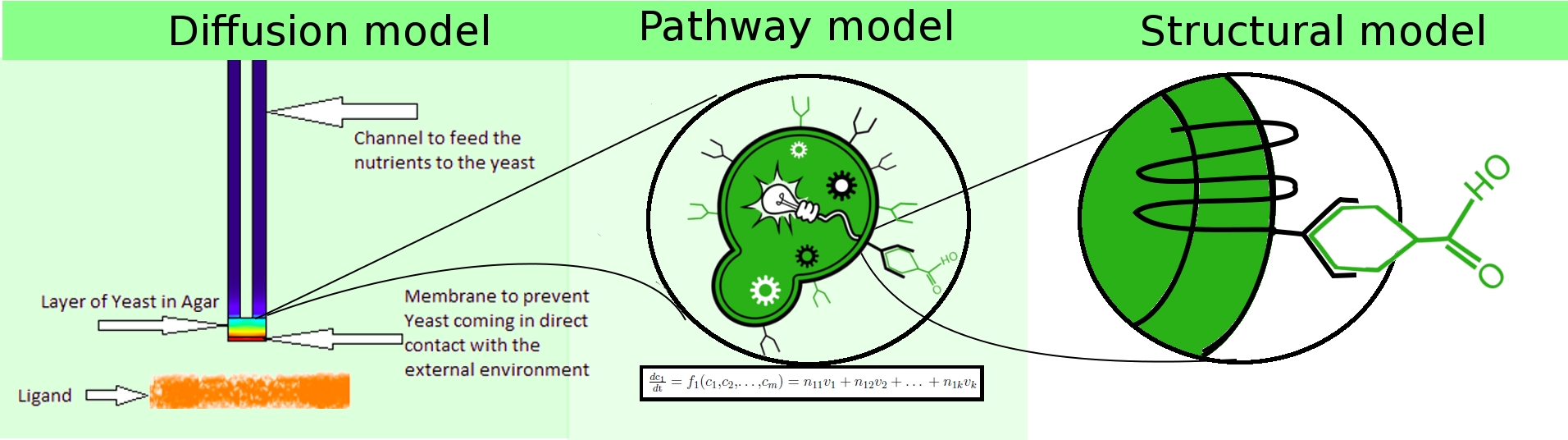
To achieve the stated objectives, we adopted a hierarchical modeling approach, in which we constructed heterogeneous models spanning
- population dynamics (diffusion model).
- extra-cellular interactions (structural model).
- intra-cellular dynamics (pathway model).
Connected Multi-level Models
These three models interact with each other to represent the mechanism of our project in different perspectives. At the system level, diffusion model sheds light on the behaviour of the proposed device the Snifferometer, incorporating the observations obtained from the single cell pathway model. At a lower level, the pathway model is built to functionally simulate the intra-cellular biochemical reactions in the signalling pathway. Parameters in pathway were fitted to the experimental data so as to provide a good estimate of the concentration of the Green Fluoroscent Protein. At a still lower level, the structural model was designed to understand the ligand docking at the molecular level. It indicates how well the ligand-receptor interaction is due to hydrogen bonds, which gives an insight into the receptor activation rate in the pathway model. Further research on the ligand-receptor model may provide more knowledge about ligand affinity and G-protein release.
Based on these models, a practical device, Snifferometer is designed to achieve the purpose of odor detection. The model of this device is developed to give an idea of the application of our project.
The models that were built spanned a broad spectrum of techniques: PDE, MD (Molecular Dynamics), SDE and ODE, as well as alignment techniques, with diverse tools being employed for the application of these techniques: COMSOL, YASARA, MATLAB, COPASI, BLAST, ClustalO, Ensembl, PSIPRED.
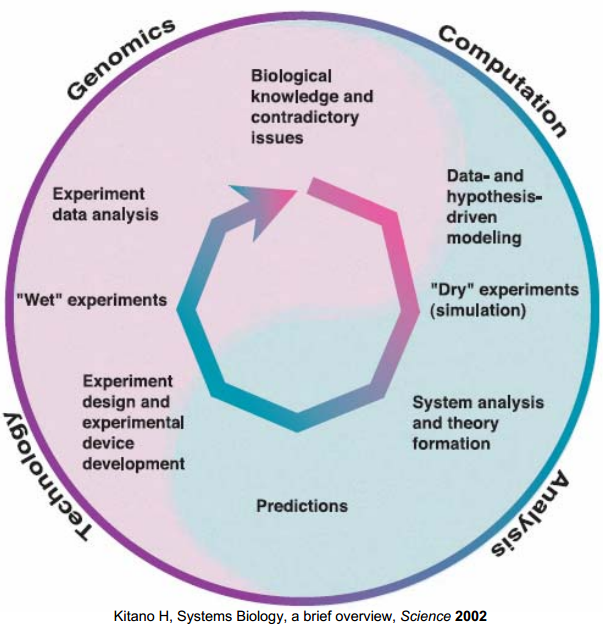
Research Cycle
This research cycle is introduced in our project as an interactive boundary between the wetlab and the "dry" modeling to gain a hierarchical in-depth understanding of our biological system. [1]
Four basic phases are sequenced to close the cycle, as shows in Figure 2.
First, model hypotheses for the signal transduction and the ligand-binding structure are proposed based on the biological expertise, the literature surveys on the related databases.
Second, due to the inadequate descriptions of dynamics and lack of information, more analysis of the hypothesized models were done to investigate the properties of models such as the parameter sensitivities and the structural stability in pathway model.
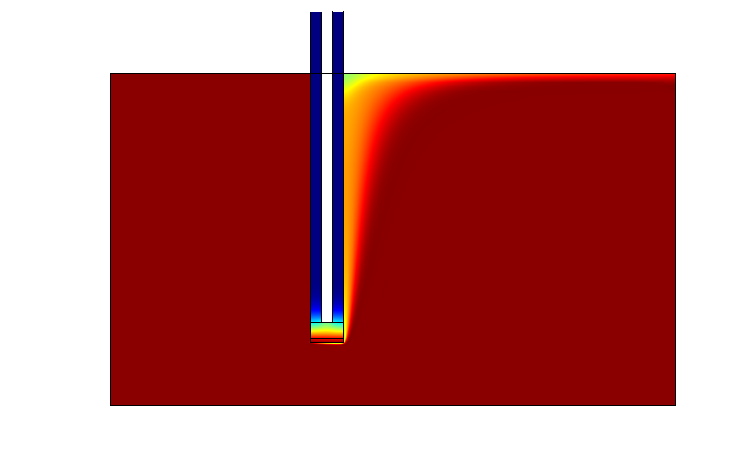
Third, The experimental design was aided by the initial analysis of the models. A 2D spatio-temporal model of the proposed device was also implemented to aid the conceptualization of the Snifferometer.
Finally, the measured data from "wet" experiment, in turn, modified the final set of parameters to provide a more accurate description of the pathway.
As a result of the research cycle, a model validated by data was obtained which was used to predict the outcomes of the experiments, using which different conditions could be simulated by "cheap" computations in silco instead of the time-consuming experiments.
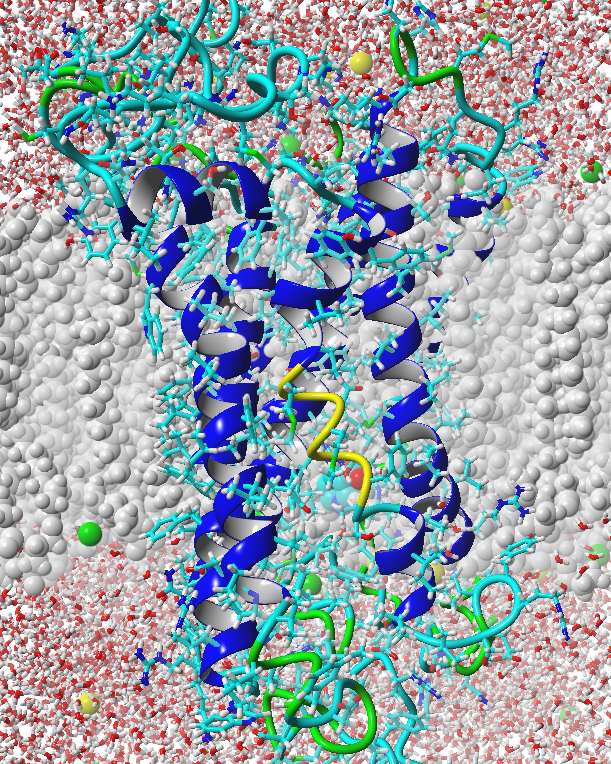
Structural Model
In order to engineer a yeast strain that is able to detect a tuberculosis molecule (TB), its receptor should be designed in such a way that the molecule can act as an agonist. By modeling the olfactory receptor in silico by means of Molecular Dynamics simulations (MD), its biophysical and biochemical properties are investigated at the molecular level. The aim is to get a clear understanding of how a ligand binds in the receptor and how to mutate the binding niche to let it bind to methyl nicotinate more specifically. A strong correlation between in vivo activity of the Homo Sapiens niacin receptor 1 and the simulated activity from the model resulted, enabling the prediction of ligand characteristics in the binding niche.
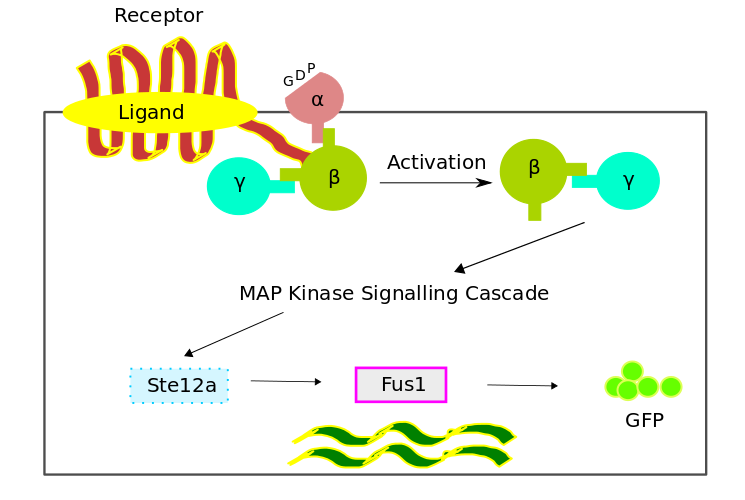
Pathway Model
The bio-chemical pathway model was developed based on a scheme favouring the temporal order of the processes, which involved the four fundamental modules,- Receptor Activation
- G - Protein Cycle
- MAPK Cascade
- Gene Expression
Three different models were used for the analysis of different aspects of the pathway. The dynamics of the system in these models were described using a set of differential equations governing the concentration changes of individual components and of complexes over time.
Sensitivity and stability analysis were permformed to determine the sensitivity coefficients which were used to study the parametric dependence of the biological models. The results from the sensitivity analysis were then used in the parameter estimation to fit the model to the data provided by the experimentalists.
Diffusion Model
One of the main objectives of the project was to synthesize a practical device for the detector This led to the design of a three layered device, The Snifferometer.
As a first step towards achieving this goal, we built a temporal model of the system using PDE's which were simulated in matlab. A 2D reaction-diffusion system was then implemented in COMSOL multiphysics using the knowledge obtained from single cell pathway model, combining the behaviours of the which helped us get a better understanding of how such a device could be implemented and the response times involved in such a process.
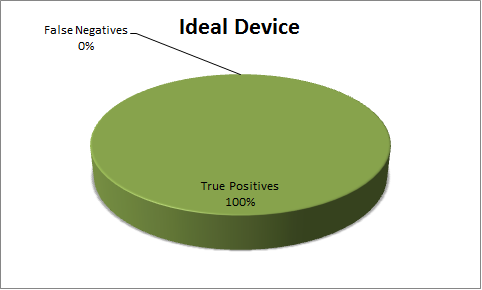
Stochastic Model - Sensitivity & Specificity Analysis
A false positive and false negative analysis is a crucial aspect of a binary classification test such as the snifferomenter, as it indicates the confidence level in the data that one should typically strive for. On account of the inherent stochasticity in biological functions, we built a stochastic model to analyze the Sensitivity and the Specificity of the Snifferometer.
Information processing Model
Signaling pathways and genetic circuitry have the capacity to transmit and process information about certain states in the environment. They are used by the cell to make decisions about whether to take certain actions to remain well adapted. Until now we have used models to describe these dynamics with the goal of eventually having enough insight into the systems so we can actually engineer them.
If we however want to quantify the information processing capacity we will need something extra. Something like the BioBit!
 "
"