Team:NTU-Taida/Modeling/2D-3D-Combined
From 2012.igem.org
(→3D Cell population response model) |
(→GLP1) |
||
| (30 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
== Overview == | == Overview == | ||
<p style="text-indent: 2em;"> | <p style="text-indent: 2em;"> | ||
| - | With the single cell model, fatty acid reaction-absorption model, and system analysis, we have verified the filter function of our circuit, simulated the real physiological fatty acid level at intestinal wall, and adjust our filter threshold to fit to the real condition. Next, we want to combine them together and see what would happen when we put cells in the intestine after a fatty meal. | + | With the single cell model, fatty acid reaction-absorption model, and system analysis, we have verified the '''filter function''' of our circuit, simulated the '''real physiological fatty acid level at intestinal wall''', and '''adjust our filter threshold to fit to the real condition'''. Next, we want to combine them together and see what would happen when we put cells in the intestine after a fatty meal. |
</p> | </p> | ||
<p style="text-indent: 2em;"> | <p style="text-indent: 2em;"> | ||
| - | The combined model is composed of three different sections. 2D combined model simulates the GLP1 response of Pepdex E-coli cells after the food intake in the x-y cross-sectional plane. 2D Cell population response model further considers the possible uneven concentration of fatty acid along the z-axis due to non-uniform distribution of lipase and simulates how the quorum sensing system assists our system to overcame this possible limitation and enhance the response. Finally, 3D Cell population response model incorporates all the consideration to generate a full model to visualize the dynamic behavior of our system. | + | The combined model is composed of three different sections. 2D combined model simulates the GLP1 response of Pepdex E-coli cells after the food intake in the x-y cross-sectional plane. 2D Cell population response model further considers the possible uneven concentration of fatty acid along the z-axis due to non-uniform distribution of lipase and simulates how the quorum sensing system assists our system to overcame this possible limitation and enhance the response. Finally, '''3D Cell population response model incorporates all the consideration to generate a full model to visualize the dynamic behavior of our system.''' |
</p> | </p> | ||
| - | |||
==2D Combined Model== | ==2D Combined Model== | ||
| Line 84: | Line 83: | ||
===Results=== | ===Results=== | ||
| - | |||
| - | |||
{| align='center' | {| align='center' | ||
| - | | {{:Team:NTU-Taida/Templates/Vid|num=3 | + | | {{:Team:NTU-Taida/Templates/Vid|num=1.1 Fatty acid concentration|width=350px|name=NTU-Taida-Model-Combined-Image5.gif}} |
| - | | {{:Team:NTU-Taida/Templates/Vid|num=3 | + | | {{:Team:NTU-Taida/Templates/Vid|num=1.2 Closer look of fatty acid concentration|width=350px|name=NTU-Taida-Model-Combined-Image8.gif}} |
| + | |} | ||
| + | {| align='center' | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=2.1 GLP-1 expression in system without quorum sensing|width=350px|name=NTU-Taida-Model-Combined-Image4.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=2.2 Closer look ofGLP-1 expression in system without quorum sensing|width=350px|name=NTU-Taida-Model-Combined-Image7.gif}} | ||
| + | |} | ||
| + | {| align='center' | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=3.1 GLP-1 expression in system with quorum sensing|width=350px|name=NTU-Taida-Model-Combined-Image3.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=3.2 Closer look at GLP-1 expression in system with quorum sensing|width=350px|name=NTU-Taida-Model-Combined-Image6.gif}} | ||
|} | |} | ||
| Line 119: | Line 124: | ||
$$\frac{\partial^2 FA}{\partial x^2} + \frac{\partial^2 FA}{\partial z^2} - \frac{1}{D_{FA}} \frac{\partial FA}{\partial t} = 0$$ | $$\frac{\partial^2 FA}{\partial x^2} + \frac{\partial^2 FA}{\partial z^2} - \frac{1}{D_{FA}} \frac{\partial FA}{\partial t} = 0$$ | ||
| - | {{:Team:NTU-Taida/Templates/Eq|num= | + | {{:Team:NTU-Taida/Templates/Eq|num=14|eq= |
}} | }} | ||
| Line 125: | Line 130: | ||
For the reaction term of AHL, we first discard the terms describing the diffusion into and out of the cell membrane in the ODEs of AHLi and AHLe in the single cell model. | For the reaction term of AHL, we first discard the terms describing the diffusion into and out of the cell membrane in the ODEs of AHLi and AHLe in the single cell model. | ||
| - | As previously mentioned, we assume that the diffusion AHL diffusion into and out of cells is fast. Therefore, we simply account for the diffusion of AHL across cell membrane by multiplying the intracellular AHLi terms by the relative cell density, which will be described in the next section. With these considerations, we combine the two equations of AHLi and AHLe (Eq. | + | As previously mentioned, we assume that the diffusion AHL diffusion into and out of cells is fast. Therefore, we simply account for the diffusion of AHL across cell membrane by multiplying the intracellular AHLi terms by the relative cell density, which will be described in the next section. With these considerations, we combine the two equations of AHLi and AHLe (Eq. 15 and Eq. 16) into one reaction equation (Eq. 17 ). |
| - | {{:Team:NTU-Taida/Templates/Eq|num= | + | {{:Team:NTU-Taida/Templates/Eq|num=15|eq=\frac{d AHL_i}{d t} = ks1 \cdot LuxI - ks0 \cdot AHL_i - \eta (AHL_i - AHL_e) |
}} | }} | ||
| - | {{:Team:NTU-Taida/Templates/Eq|num= | + | {{:Team:NTU-Taida/Templates/Eq|num=16|eq=\frac{d AHL_e}{d t} = - kse \cdot AHL_e + \eta_{Ext} (AHL_i - AHL_e) |
}} | }} | ||
$$R (AHL) = - \gamma_{AHL_{ext}} \cdot AHL + cd (k_{s1} \cdot LuxI - k_{s0} \cdot AHL)$$ | $$R (AHL) = - \gamma_{AHL_{ext}} \cdot AHL + cd (k_{s1} \cdot LuxI - k_{s0} \cdot AHL)$$ | ||
| - | {{:Team:NTU-Taida/Templates/Eq|num= | + | {{:Team:NTU-Taida/Templates/Eq|num=17|eq= |
}} | }} | ||
| - | With the help of COMSOL Multiphysics, we can consider both the diffusion and reaction terms of AHL by giving the diffusion (Eq. | + | With the help of COMSOL Multiphysics, we can consider both the diffusion and reaction terms of AHL by giving the diffusion (Eq. 14) and reaction (Eq. 17) equations above, without the need to solve the explicit PDE of AHL. |
The diffusion of AHL is modeled by a free diffusion equation. | The diffusion of AHL is modeled by a free diffusion equation. | ||
$$ \frac{\partial^2 AHL}{\partial x^2} + \frac{\partial^2 AHL}{\partial z^2} - \frac{1}{D_{AHL}} \frac{\partial AHL}{\partial t} = 0$$ | $$ \frac{\partial^2 AHL}{\partial x^2} + \frac{\partial^2 AHL}{\partial z^2} - \frac{1}{D_{AHL}} \frac{\partial AHL}{\partial t} = 0$$ | ||
| - | {{:Team:NTU-Taida/Templates/Eq|num= | + | {{:Team:NTU-Taida/Templates/Eq|num=18|eq= |
}} | }} | ||
| Line 166: | Line 171: | ||
====FA==== | ====FA==== | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=4|width= | + | {| align='center' |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=4.1 without quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video4-1.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=4.2 with quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video4-2.gif}} | ||
| + | |} | ||
====X_fadR==== | ====X_fadR==== | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=5|width= | + | {| align='center' |
| - | {{:Team:NTU-Taida/Templates/Vid|num=6|width= | + | | {{:Team:NTU-Taida/Templates/Vid|num=5.1 without quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video5-1.gif}} |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=5.2 with quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video5-2.gif}} | ||
| + | |} | ||
| + | |||
| + | ====TetR1==== | ||
| + | {| align='center' | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=6.1 without quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video6-1.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=6.2 with quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video6-2.gif}} | ||
| + | |} | ||
===Species regulated by the quorum sensing module=== | ===Species regulated by the quorum sensing module=== | ||
====LacI==== | ====LacI==== | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=7|width= | + | |
| - | The expression of LacI is inhibited by TetR proteins, and therefore is regulated by the quorum sensing module. Although the concentration of TetR1 is the same in systems with and without quorum sensing, cells with quorum sensing produce TetR2 as the output of the quorum sensing module and therefore result in overall more total TetR proteins. Thus, the repression of LacI is more significant in systems with the quorum sensing module. | + | {|align="center" |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=7-1 without quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video7-1.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=7-2 with quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video7-2.gif}} | ||
| + | |} | ||
| + | |||
| + | The expression of LacI is inhibited by TetR proteins, and therefore is regulated by the quorum sensing module. Although the concentration of TetR1 is the same in systems with and without quorum sensing, cells with quorum sensing produce TetR2 as the output of the quorum sensing module and therefore result in overall more total TetR proteins. Thus, the repression of LacI is more significant in systems with the quorum sensing module. | ||
====GLP1==== | ====GLP1==== | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=8|width= | + | {|align="center" |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=8-1 without quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video8-1.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=8-2 with quorum sensing |width=350px|name=NTU-Taida-Model-Combined-video8-2.gif}} | ||
| + | |} | ||
| + | |||
As the expression of GLP1 is repressed by LacI proteins, GLP1 also displays different spatial-temporal concentration pattern between the two systems. With quorum sensing module, the system produces more GLP1 overall. | As the expression of GLP1 is repressed by LacI proteins, GLP1 also displays different spatial-temporal concentration pattern between the two systems. With quorum sensing module, the system produces more GLP1 overall. | ||
===Effect of cell density=== | ===Effect of cell density=== | ||
| - | Relative cell density plays a critical role in determining whether we can see a significant difference between systems with and without quorum sensing. We simulated the spatial-temporal response of GLP1 of four systems, all of them with the quorum sensing module but with relative cell densities of 0.2, 0.1, and 0.01, respectively. | + | Relative cell density plays a critical role in determining whether we can see a significant difference between systems with and without quorum sensing. We simulated the spatial-temporal response of GLP1 of four systems, all of them with the quorum sensing module but with relative cell densities of 0.2, 0.1, 0.05, and 0.01, respectively. |
Then we compare their response to that of system without quorum sensing to determine if there exists a threshold of cell density, below which the effect of quorum sensing module is unperceivable. | Then we compare their response to that of system without quorum sensing to determine if there exists a threshold of cell density, below which the effect of quorum sensing module is unperceivable. | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=9|width= | + | {|align="center" |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=9-1 cd = 0.2 |width=280px|name=NTU-Taida-Model-Combined-video9-1.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=9-2 cd = 0.1 |width=280px|name=NTU-Taida-Model-Combined-video9-2.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=9-3 cd = 0.05 |width=280px|name=NTU-Taida-Model-Combined-video9-3.gif}} | ||
| + | |} | ||
| - | + | {|align="center" | |
| + | | {{:Team:NTU-Taida/Templates/Vid|num=9-4 cd = 0.01 |width=280px|name=NTU-Taida-Model-Combined-video9-4.gif}} | ||
| + | | {{:Team:NTU-Taida/Templates/Vid|num=9-5 without quorum sensing module |width=280px|name=NTU-Taida-Model-Combined-video9-5.gif}} | ||
| + | |} | ||
| + | |||
| + | |||
| + | We can see from Video 9 that the system with a relative cell density of 0.2 differs most with the system without the quorum sensing module. The system with a relative cell density of 0.1 shows moderate difference. The system with a relative cell density of 0.05 displays very subtle difference. There is no difference between the system with a relative cell density of 0.01 and the system with no quorum sensing module. Therefore, the threshold of relative cell density may lie between 0.05 and 0.01. | ||
==3D Cell population response model== | ==3D Cell population response model== | ||
| Line 201: | Line 236: | ||
{|align="center" | {|align="center" | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=10 Full Thumbnails |width= | + | | {{:Team:NTU-Taida/Templates/Vid|num=10 Full Thumbnails |width=400px|name=image24.gif}} |
| - | {{:Team:NTU-Taida/Templates/Vid|num=10 Enlargement |width= | + | | {{:Team:NTU-Taida/Templates/Vid|num=10 Enlargement |width=400px|name=image26.gif}} |
|} | |} | ||
{|align="center" | {|align="center" | ||
| - | {{:Team:NTU-Taida/Templates/Vid|num=11 Full Thumbnails |width= | + | | {{:Team:NTU-Taida/Templates/Vid|num=11 Full Thumbnails |width=400px|name=image25.gif}} |
| - | {{:Team:NTU-Taida/Templates/Vid|num=11 Enlargement |width= | + | | {{:Team:NTU-Taida/Templates/Vid|num=11 Enlargement |width=400px|name=image27.gif}} |
|} | |} | ||
Latest revision as of 14:59, 3 January 2013
2D & 3D Combined Model
Contents |
Overview
With the single cell model, fatty acid reaction-absorption model, and system analysis, we have verified the filter function of our circuit, simulated the real physiological fatty acid level at intestinal wall, and adjust our filter threshold to fit to the real condition. Next, we want to combine them together and see what would happen when we put cells in the intestine after a fatty meal.
The combined model is composed of three different sections. 2D combined model simulates the GLP1 response of Pepdex E-coli cells after the food intake in the x-y cross-sectional plane. 2D Cell population response model further considers the possible uneven concentration of fatty acid along the z-axis due to non-uniform distribution of lipase and simulates how the quorum sensing system assists our system to overcame this possible limitation and enhance the response. Finally, 3D Cell population response model incorporates all the consideration to generate a full model to visualize the dynamic behavior of our system.
2D Combined Model
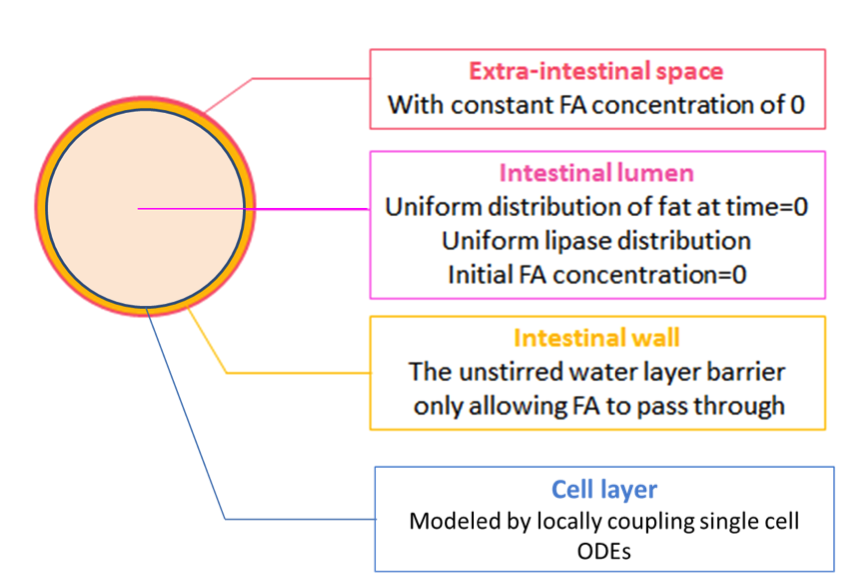
So far, we have the spatial-temporal concentration of fatty acid, and cells with suitable threshold. In this model, we want to see what would happen when we put cells in the intestine after a meal.
We achieve this by further coupling the single cell ODEs (with parameters adjusted by system analysis) locally to the fatty acid reaction-absorption model at the intestinal wall, as shown in Figure 2.
Besides to see the cell response when combined with the environmental input, we also want to simulate the prolonged effect of the quorum sensing module with physiological input. Therefore, we simulate system with and without quorum sensing modules and compare their results.
Spatial-temporal Model Equations
The equations used in our 2D combined model mainly come from the reaction-absorption model and the single cell model. The following three equations are the same as those in the reaction-absorption model, which simulates the change in fatty acid concentration in time and space.
$$\frac{d fat}{d t} = -\frac{k_{cat}[E]_t\cdot[S]}{K_m + [S]}$$
$$\frac{d FA}{d t} = 3 \cdot \frac{k_{cat}[E]_t\cdot[S]}{K_m + [S]}$$
$$J=(C_1 - C_2)(D_{FA}/d)$$
Other equations in the model are almost the same as those derived in our single cell model, with slightly difference in those for AHL, which will be discussed thoroughly in the 2D Cell population response model. The equations are shown as below.
$$\frac{d FadR}{d t} = \alpha_{FadR} - \gamma_{FadR} \cdot FadR$$
$$X_{FadR} = \frac{FadR \cdot \beta_{FA}^{n2}}{FA^{n2} + \beta_{FA}^{n2}}$$
$$$$
$$\frac{dTetR_1}{dt} = \frac{\alpha_{TetR_1} \cdot {\beta_{FadR}^{n1}}}{\beta_{FadR}^{n1} + X_{FadR}^{n1}} - \gamma_{TetR_1} \cdot TetR_1 $$
$$$$
$$\frac{d TetR_2}{d t} = \frac{\alpha_{TetR_2} \cdot R^{n3}}{\beta_R^{n3} + R^{n3}} - \gamma_{TetR_2} \cdot TetR_2$$
$$$$
$$\frac{d LuxI}{d t} = \frac{\alpha_{LuxI} \cdot \beta_{FadR}^{n1}}{\beta_{FadR}^{n1} + X_{FadR}^{n1}} - \gamma_{LuxI} \cdot LuxI$$
$$$$
$$\frac{\partial^2 AHL}{\partial x^2} + \frac{\partial^2 AHL}{y^2} - \frac{1}{D_{AHL}}\cdot \frac{\partial AHL}{\partial t} = 0$$
$$$$
$$R(AHL) = - \gamma_{AHL_{ext}} \cdot AHL + cd(k_{s1}LuxI - k_{s0}AHL)$$
$$$$
$$\frac{d R}{d t} = \rho (LuxR)^2(AHL_i)^2 - \gamma_R \cdot R$$
$$$$
$$\frac{d LacI}{d t} = \frac{\alpha_{LacI}}{1 + (\frac{TetR_1 + TetR_2}{\beta_{TetR}})^{n5}} - \gamma_{LacI} \cdot LacI$$
$$$$
$$\frac{d GLP1}{d t} = \frac{\alpha_{GLP1}}{1 + (\frac{LacI}{\beta_{LacI}})^{n6}} - \gamma_{GLP1} \cdot GLP1$$
$$$$
For the control group without the quorum sensing module, we simply discard the terms related to quorum sensing system in the ODE set.
Results
Video 1 shows the spatial temporal concentration change of fatty acid. Video 2 and three show the corresponding spatial temporal response of GLP-1 expression in system without and with quorum sensing module, respectively.
We can see from the videos that the fatty acid concentration at the intestinal wall first increases rapidly, turning on the circuits in the cells and resulting in GLP-1 expression at the intestinal wall. Then due to the absorption of fatty acid, the concentration gradually falls and the GLP-1 concentration started to decrease. Notice that the GLP1 expression in the system with Quorum sensing, shown in Video 3, lasts longer than that of the system without quorum sensing, shown in Video 2. Consistent to the single cell model, the system with QS has more prolonged response compared to the one without it.
Cell Population Response Model
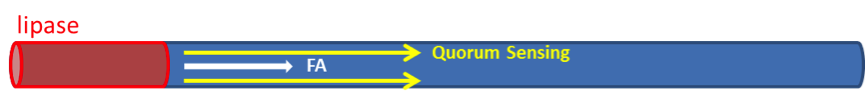
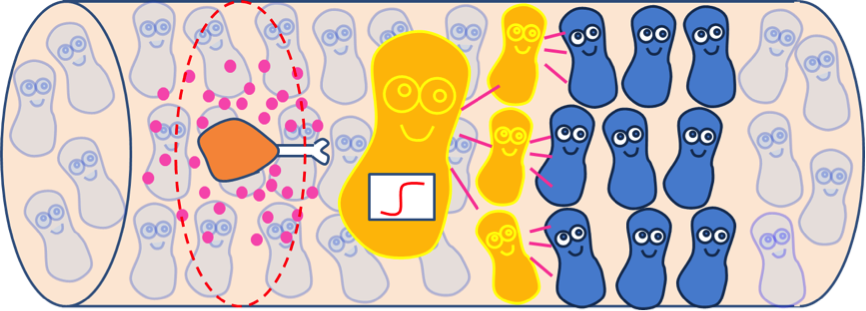
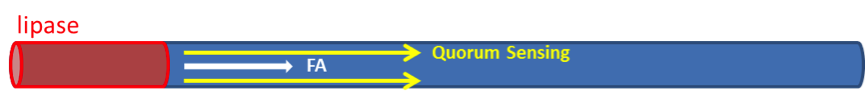
We have assumed that fat distributes uniformly along the z-axis in the lumen after a meal in the previous models. However, in real situation in human body, lipase only exists in certain segments of the intestine. Therefore, fat hydrolysis will only take place in those segments and there will be no fatty acid produced outside these segments, as shown in Figure 3. Cells residing in the segments without lipase activity can only sense the fatty acid diffused from the source segments. Because AHL diffuses faster than fatty acid, we incorporate the quorum sensing system to enable faster recruitment of more bacteria to express GLP-1 in regions without lipase.
Quorum sensing module enables cells to communicate with each other, as the uneven distribution of fatty acid in the intestines may cause gaps in individual detection. Cells with the quorum sensing module can respond to food intake event as long as their neighboring cells have detected fatty acid, enhancing the overall response to the food intake (Figure 4). However, the communication between cells via AHL can’t be seen when only viewing a single cell. Therefore, we construct spatial temporal models with COMSOL Multiphysics in order to gain insights into the influence of cell-cell communication mediated via the quorum sensing module on the overall GLP1 expression.
2D Cell Population Response Model
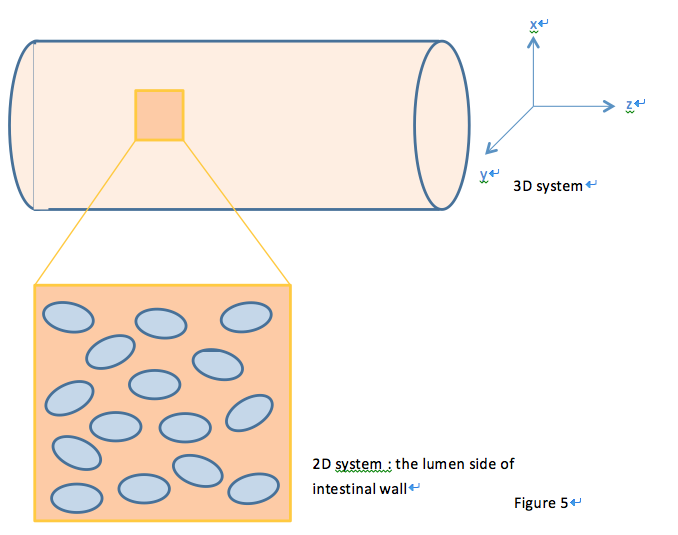
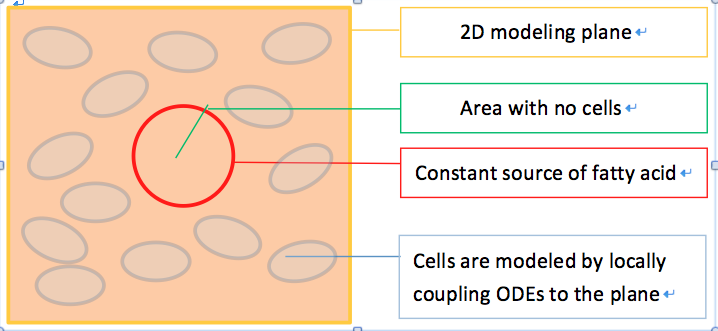
We start our effort with a 2D spatial temporal model. When E.coli cells colonize the intestine, they tend to attach onto the intestinal walls instead of distributing evenly throughout the intestine lumen. Therefore, we first view our system as a two dimensional system, with the modeling plane in the x-z (or either y-z) dimension.
Geometry Design
With this in mind, we construct a plane in 2D geometry and couple the ODEs derived in our single cell model to simulate the presence of cells.
To model the effect of quorum sensing, we establish an uneven distribution of fatty acid by giving a constant concentration source of fatty acid in the middle of our modeled plane, as shown in Figure 6. There are no cells within the circular fatty acid source. As time goes on, fatty acids diffuse from the source and establish a concentric gradient. To see the effect of quorum sensing mechanism on overall GLP1 expression in a cell population with uneven distributed fatty acid input, we model cells with and without quorum sensing and compare their spatial –temporal GLP1 expression.
Spatial-temporal Model Equations
The first equation used in our model describes the free diffusion of fatty acids from the central source.
$$\frac{\partial^2 FA}{\partial x^2} + \frac{\partial^2 FA}{\partial z^2} - \frac{1}{D_{FA}} \frac{\partial FA}{\partial t} = 0$$
$$$$
Other equations in the model are almost the same as those derived in our single cell model, except for the ones for AHL. For AHL, we have to set up a partial differential equation describing both the diffusion and the reaction event of AHL. We assume that AHL diffusion into and out of cells is fast and do not model this diffusion process explicitly. Therefore, in contrast to the single-cell model, we only use one species called AHL instead of an internal AHL concentration AHLi and an external AHL concentration AHLe.
For the reaction term of AHL, we first discard the terms describing the diffusion into and out of the cell membrane in the ODEs of AHLi and AHLe in the single cell model. As previously mentioned, we assume that the diffusion AHL diffusion into and out of cells is fast. Therefore, we simply account for the diffusion of AHL across cell membrane by multiplying the intracellular AHLi terms by the relative cell density, which will be described in the next section. With these considerations, we combine the two equations of AHLi and AHLe (Eq. 15 and Eq. 16) into one reaction equation (Eq. 17 ).
$$\frac{d AHL_i}{d t} = ks1 \cdot LuxI - ks0 \cdot AHL_i - \eta (AHL_i - AHL_e)$$
$$\frac{d AHL_e}{d t} = - kse \cdot AHL_e + \eta_{Ext} (AHL_i - AHL_e)$$
$$R (AHL) = - \gamma_{AHL_{ext}} \cdot AHL + cd (k_{s1} \cdot LuxI - k_{s0} \cdot AHL)$$
$$$$
With the help of COMSOL Multiphysics, we can consider both the diffusion and reaction terms of AHL by giving the diffusion (Eq. 14) and reaction (Eq. 17) equations above, without the need to solve the explicit PDE of AHL.
The diffusion of AHL is modeled by a free diffusion equation.
$$ \frac{\partial^2 AHL}{\partial x^2} + \frac{\partial^2 AHL}{\partial z^2} - \frac{1}{D_{AHL}} \frac{\partial AHL}{\partial t} = 0$$
$$$$
Other equations in the model are the same as those derived in our single cell model. For the control group without the quorum sensing module, we discard the terms related to quorum sensing system in the ODE set.
Relative Cell density
The relative cell density is defined as the ratio of the total area E.coli cells occupy to the area of the intestinal wall. By multiplying the intracellular AHL synthesis and degradation terms by the relative cell density, we assume that as a cell synthesizes or degrades AHL, the addition or reduction of AHL molecules is dispersed evenly through the neighboring area. (Mind that the diffusion of these AHL molecules on global scale is still governed by the free diffusion equation.)
We calculated the relative cell density by assuming there are 1014 bacteria cells residing in the intestine, which has a total surface area of 200 m2. Thus, the average cell density is 5*1011 cells/m2. We can measure the surface area of each individual cell by multiplying their length by their width, giving a surface area of 10-12 m2 per cell. Multiplying the surface area per cell and the average cell density gives the area density of total cells as 0.5. Since 99% of the gut flora consists of 30-40 species of bacteria, we can assume that each of the common types contribute to approximately 1% of the total surface area of the intestine. Considering the clustering effect of cells, we assume that some regions would have much higher cell densities than others, such that the uneven distribution may result in a tenfold fluctuation in cell density. We hypothesize that the relative cell density in our model could be as high as 0.1.
Cell density has dramatic effect on the overall effect of quorum sensing system, as supported by a recent study investigating the synchronization of repressilators by quorum sensing mechanisms [1]. Therefore, we also model our system with different cell densities, as can be seen in the results.
Results
To see the effect of quorum sensing mechanism on overall GLP1 expression in a cell population with uneven distributed fatty acid input, we model cells with and without quorum sensing and compare their spatial - temporal GLP1 expression, with relative cell density of 0.1 The results show that species which are not regulated by the quorum sensing module, such as FA, X_FadR, and TetR1, show no difference in the spatial temporal expression pattern between system with and without the quorum sensing system.
However, species affected by the quorum sensing module, such as LacI and GLP1, show striking difference in their spatial temporal concentration pattern. Within the given time span, systems with a quorum sensing module produces GLP1 over a broader spatial region, resulting in more total GLP1.
Species unregulated by the quorum sensing module
FA
X_fadR
TetR1
Species regulated by the quorum sensing module
LacI
The expression of LacI is inhibited by TetR proteins, and therefore is regulated by the quorum sensing module. Although the concentration of TetR1 is the same in systems with and without quorum sensing, cells with quorum sensing produce TetR2 as the output of the quorum sensing module and therefore result in overall more total TetR proteins. Thus, the repression of LacI is more significant in systems with the quorum sensing module.
GLP1
As the expression of GLP1 is repressed by LacI proteins, GLP1 also displays different spatial-temporal concentration pattern between the two systems. With quorum sensing module, the system produces more GLP1 overall.
Effect of cell density
Relative cell density plays a critical role in determining whether we can see a significant difference between systems with and without quorum sensing. We simulated the spatial-temporal response of GLP1 of four systems, all of them with the quorum sensing module but with relative cell densities of 0.2, 0.1, 0.05, and 0.01, respectively.
Then we compare their response to that of system without quorum sensing to determine if there exists a threshold of cell density, below which the effect of quorum sensing module is unperceivable.
We can see from Video 9 that the system with a relative cell density of 0.2 differs most with the system without the quorum sensing module. The system with a relative cell density of 0.1 shows moderate difference. The system with a relative cell density of 0.05 displays very subtle difference. There is no difference between the system with a relative cell density of 0.01 and the system with no quorum sensing module. Therefore, the threshold of relative cell density may lie between 0.05 and 0.01.
3D Cell population response model
Finally, we construct a full 3D model, combing the consideration on the x-y plane and the x-z plane. Our goal is to simulate the effect of non-uniform production of fatty acid due to the spatially limited lipase activity. We modeled the intestine by a cylindrical tube, with a fat source limited to the z=0 plane, to simulate the situations similar to that shown in Figure 7, and examine the effect of Quorum sensing.
The equations used are the same as those in the 2D cell population response model, but with the equations governing diffusions changing from the two-dimension form to their three-dimension counterparts.
Video 10 shows the result of system without quorum sensing module while Video 11 shows the result of system with quorum sensing module. From Video 10 and 11, we can see that GLP-1 starts to be expressed at similar time point at the intestinal wall on the z=0 plane. However, the GLP-1 expression spreads faster along the z-axis in system with quorum sensing.
Reference
- Jordi Garcia-Ojalvo , Michael B. Elowitz, and Steven H. Strogatz , Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing, pnas,2004
 "
"