Team:TU-Delft/Modeling/StochasticSensitivitySpecificityAnalysis
From 2012.igem.org
| (17 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
<center> | <center> | ||
| - | <img src="https://static.igem.org/mediawiki/ | + | <img src="https://static.igem.org/mediawiki/2012/3/30/StochasticPageHeader.jpg" align="middle" width="100%"> |
</center> | </center> | ||
| Line 17: | Line 17: | ||
<div id="contentbox" style="text-align:justify;"> | <div id="contentbox" style="text-align:justify;"> | ||
Biological functions are inherently stochastic in nature, which leads to a wide degree of variability not only at the population level but also at the level of individual cells, which makes it important to test the reliability of our system. Towards this, we first built a stochastic model of the pathway and then used it to assess the specificity and the sensitivity of our device. | Biological functions are inherently stochastic in nature, which leads to a wide degree of variability not only at the population level but also at the level of individual cells, which makes it important to test the reliability of our system. Towards this, we first built a stochastic model of the pathway and then used it to assess the specificity and the sensitivity of our device. | ||
| - | |||
| - | |||
| - | |||
__TOC__ | __TOC__ | ||
= Stochastic Model = | = Stochastic Model = | ||
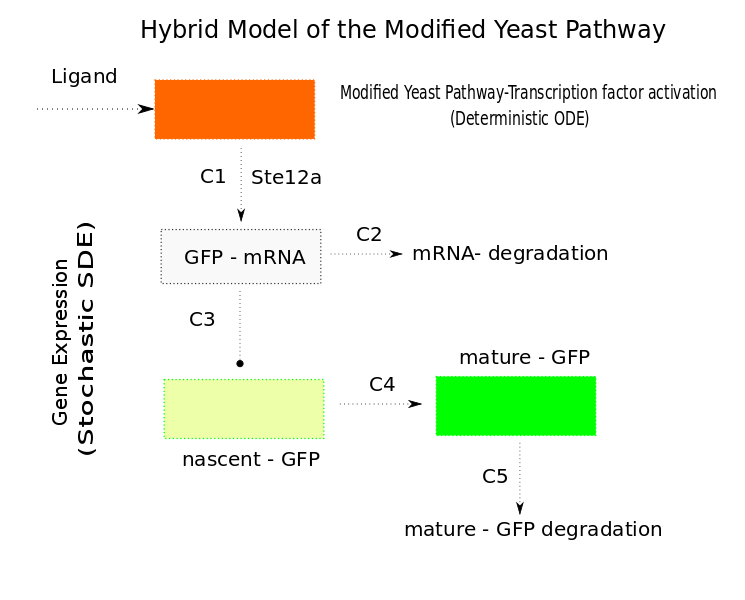
| - | In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population [[Team:TU-Delft/Modeling/SingleCellModel# | + | In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population [[Team:TU-Delft/Modeling/SingleCellModel#Ref1|[1]]].[[File:TUDHybridModel.png|400px|left|thumb|'''Figure 1''': Hybrid model of the modified yeast pathway]] In order to investigate the effects of this stochasticity, we decided to build a stochastic model of the pathway using a '''Hybrid ODE-SDE''' framework (where SDE stands for stochastic differential equation). Motivated by the fact that the Pheromone signalling is robust against cell to cell variations., we assume the dynamics until the activation of the transcription factor to be deterministic rather than stochastic. As a result of the gene expression being noisy, we build a hybrid stochastic model consisting of deterministic semantics until the activation of the transcription factor ''Ste12'' and treat it as a time varying parameter modulating the reaction based gene expression module interpreted with stochastic semantics using the stochastic differential equations approach [[Team:TU-Delft/Modeling/SingleCellModel#Ref2|[2]]], the schematic of which is given in Figure 1. |
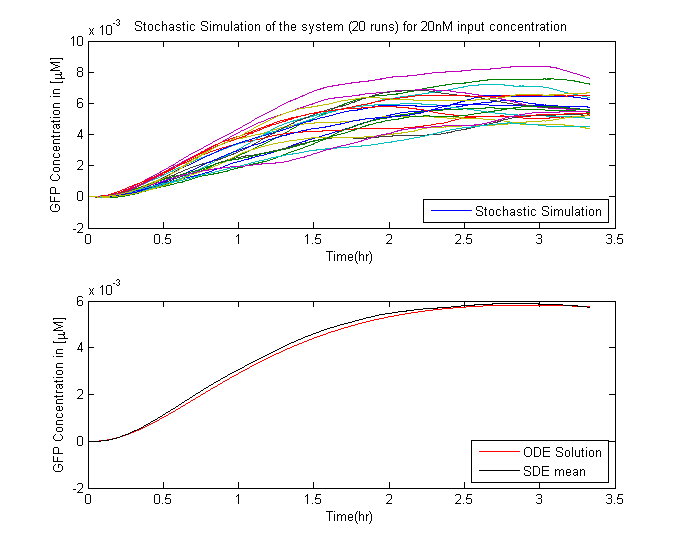
| - | Twenty simulations were done for | + | Twenty simulations were done for different input concentrations. The results of which are in the figure below. |
| - | [[File:CLE20nM.png|575px|left|thumb|'''Figure | + | [[File:CLE20nM.png|575px|left|thumb|'''Figure 2''': Stochastic Simulation of the System for 20nM Ligand concentration]] |
| - | [[File:CLE200nM.png|575px|left|thumb|'''Figure | + | [[File:CLE200nM.png|575px|left|thumb|'''Figure 3''': Stochastic Simulation of the System for 200nM Ligand concentration]] |
</div> | </div> | ||
<div id="contentbox" style="text-align:justify;"> | <div id="contentbox" style="text-align:justify;"> | ||
| - | = | + | = Sensitivity and Specificity = |
| - | + | ||
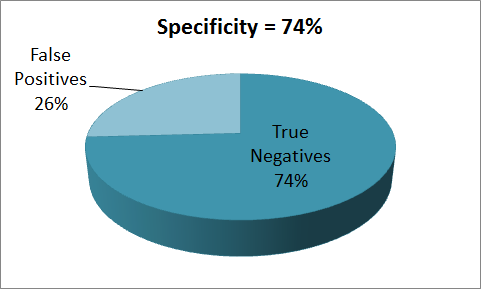
| - | + | Using an input ligand concentration of 200uM (this corresponds to the quantity of ''Methyl Nicotinate'' in TB sample), we used the single cell pathway model to set the threshold to the maximum GFP output that was produced. We then used a sample population of 1000 people and a input ligand concentration varying over a range from 0 - 2uM to perform the test. The results from the tests are presented below. | |
| - | + | ||
| + | = Results = | ||
| + | |||
| + | [[File:FN.png|260px|left|thumb|'''Figure 4''': False Negative Analysis]] | ||
| + | [[File:FP.png|260px|right|thumb|'''Figure 5''': False Positive Analysis]] | ||
| + | |||
| + | = Conclusion = | ||
| + | |||
| + | The results indicate that the proposed device has a sensitivity of 87% and a specificity of 74%, which shows that there is a great potential for further improvement. Towards which we are currently making use of the structural model to investigate ways by which better ligand binding affinities can be achieved. | ||
| + | |||
| + | = MATLAB Codes = | ||
| + | === Stochastic Model & False positive;False negative tests === | ||
| + | [[File:TUD-Download.png|50px|link=File:FalsePositiveFalseNegativeTest.zip|left]] | ||
| + | <br clear="all" /> | ||
| + | |||
| + | = References = | ||
| + | |||
| + | {|style="margin: 1em auto 1em auto;" class="wikitable sortable" width="90%" | ||
| + | !style="white-space: nowrap; width: 1px;"| | ||
| + | !class="unsortable"|Source | ||
| + | |- | ||
| + | |style="text-align: center"|<span id="Ref1">[1]</span> | ||
| + | |[http://www.nature.com/ng/journal/v31/n1/pdf/ng869.pdf Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, <br>'''Regulation of noise in the expression of a single gene''', <br>Nature Genet. 31, 69-73 (2002)] | ||
| + | |- | ||
| + | |style="text-align: center"|<span id="Ref2">[2]</span> | ||
| + | |[http://epubs.siam.org/doi/pdf/10.1137/060666457 Desmond J.Hingham<br>'''Modeling and Simulating Chemical Reactions''',<br>SIAM Rev., 50(2), 347–368] | ||
| + | |} | ||
| + | |||
| + | </div> | ||
| + | |||
| + | <div> | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/igem.org/3/37/Footer_2.jpg" align="middle" width="690"></img> | ||
| + | <a href='https://2012.igem.org/Main_Page' target="_blank"><div id='logo_igem2'></div><a/> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | </div> | ||
Latest revision as of 03:38, 27 October 2012


Biological functions are inherently stochastic in nature, which leads to a wide degree of variability not only at the population level but also at the level of individual cells, which makes it important to test the reliability of our system. Towards this, we first built a stochastic model of the pathway and then used it to assess the specificity and the sensitivity of our device.
Contents |
Stochastic Model
In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population [1]. In order to investigate the effects of this stochasticity, we decided to build a stochastic model of the pathway using a Hybrid ODE-SDE framework (where SDE stands for stochastic differential equation). Motivated by the fact that the Pheromone signalling is robust against cell to cell variations., we assume the dynamics until the activation of the transcription factor to be deterministic rather than stochastic. As a result of the gene expression being noisy, we build a hybrid stochastic model consisting of deterministic semantics until the activation of the transcription factor Ste12 and treat it as a time varying parameter modulating the reaction based gene expression module interpreted with stochastic semantics using the stochastic differential equations approach [2], the schematic of which is given in Figure 1.Twenty simulations were done for different input concentrations. The results of which are in the figure below.
Sensitivity and Specificity
Using an input ligand concentration of 200uM (this corresponds to the quantity of Methyl Nicotinate in TB sample), we used the single cell pathway model to set the threshold to the maximum GFP output that was produced. We then used a sample population of 1000 people and a input ligand concentration varying over a range from 0 - 2uM to perform the test. The results from the tests are presented below.
Results
Conclusion
The results indicate that the proposed device has a sensitivity of 87% and a specificity of 74%, which shows that there is a great potential for further improvement. Towards which we are currently making use of the structural model to investigate ways by which better ligand binding affinities can be achieved.
MATLAB Codes
Stochastic Model & False positive;False negative tests
References
| Source | |
|---|---|
| [1] | [http://www.nature.com/ng/journal/v31/n1/pdf/ng869.pdf Ozbudak, E. M., Thattai, M., Kurtser, I., Grossman, A. D. & van Oudenaarden, Regulation of noise in the expression of a single gene, Nature Genet. 31, 69-73 (2002)] |
| [2] | [http://epubs.siam.org/doi/pdf/10.1137/060666457 Desmond J.Hingham Modeling and Simulating Chemical Reactions, SIAM Rev., 50(2), 347–368] |
 "
"