Team:Bielefeld-Germany/Amsterdam/Labjournal
From 2012.igem.org
(→Monday October 22nd) |
JSchirmacher (Talk | contribs) (→Monday October 22nd) |
||
| (117 intermediate revisions not shown) | |||
| Line 119: | Line 119: | ||
**** 7/16 positive | **** 7/16 positive | ||
**** plated three on select-agar | **** plated three on select-agar | ||
| + | |||
* '''Team Shuttle Vector:''' | * '''Team Shuttle Vector:''' | ||
| - | ** The induction with 0.5% (v/v) methanol of P. pastoris GS115 cells with integrated | + | ** The induction with 0.5% (v/v) methanol of P. pastoris GS115 cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] was started. |
| + | |||
* '''Team Substrate Analysis''': | * '''Team Substrate Analysis''': | ||
** Since we have seen some new peaks after Estradiol treated with Laccases in the LC-MS, we wanted to have more degradation products to do an MS so we used a higher concentradion of Estradiol and Laccase and let them incubate for 66 hours. | ** Since we have seen some new peaks after Estradiol treated with Laccases in the LC-MS, we wanted to have more degradation products to do an MS so we used a higher concentradion of Estradiol and Laccase and let them incubate for 66 hours. | ||
| Line 126: | Line 128: | ||
* '''Team Cultivation and Purification''' | * '''Team Cultivation and Purification''' | ||
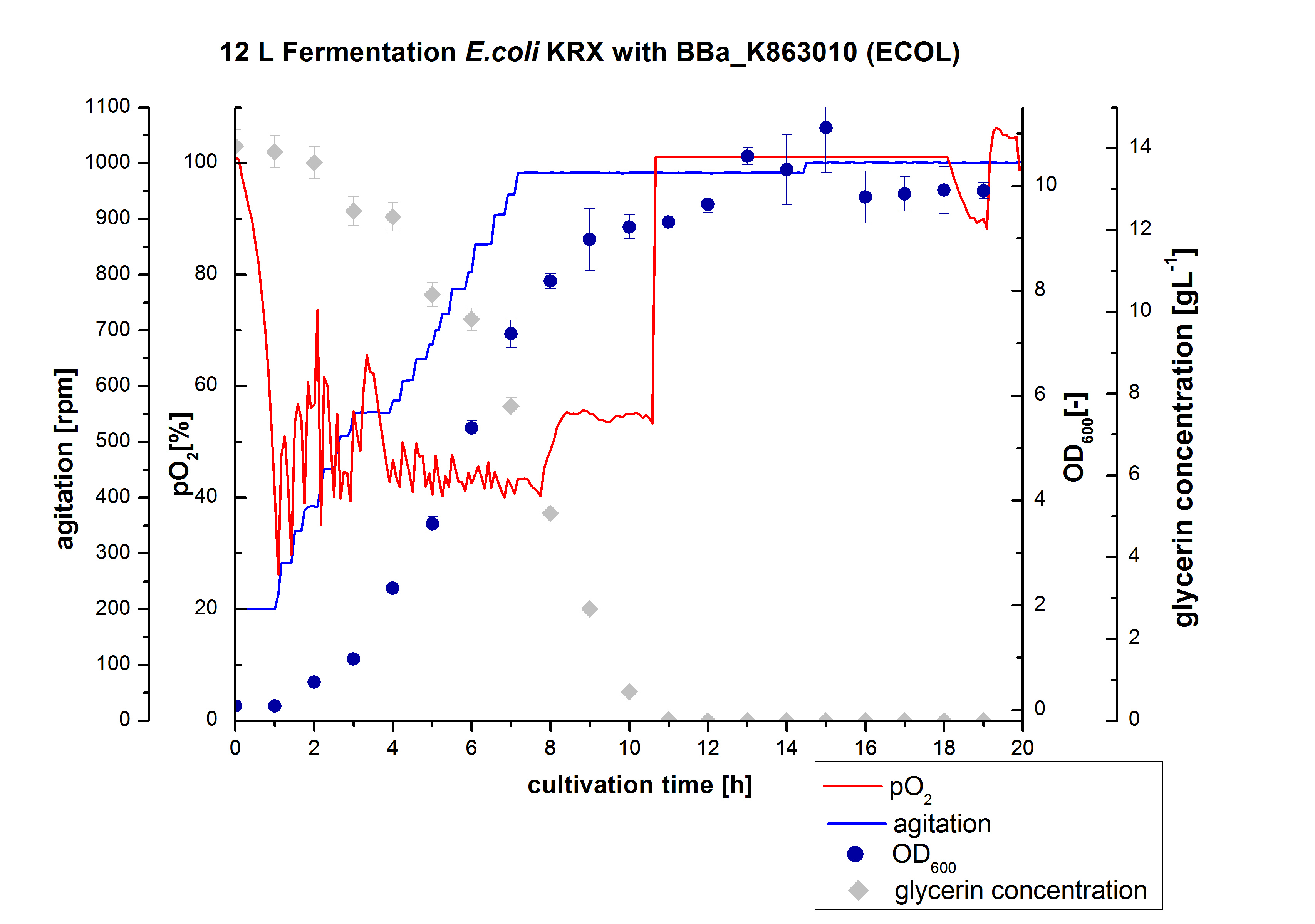
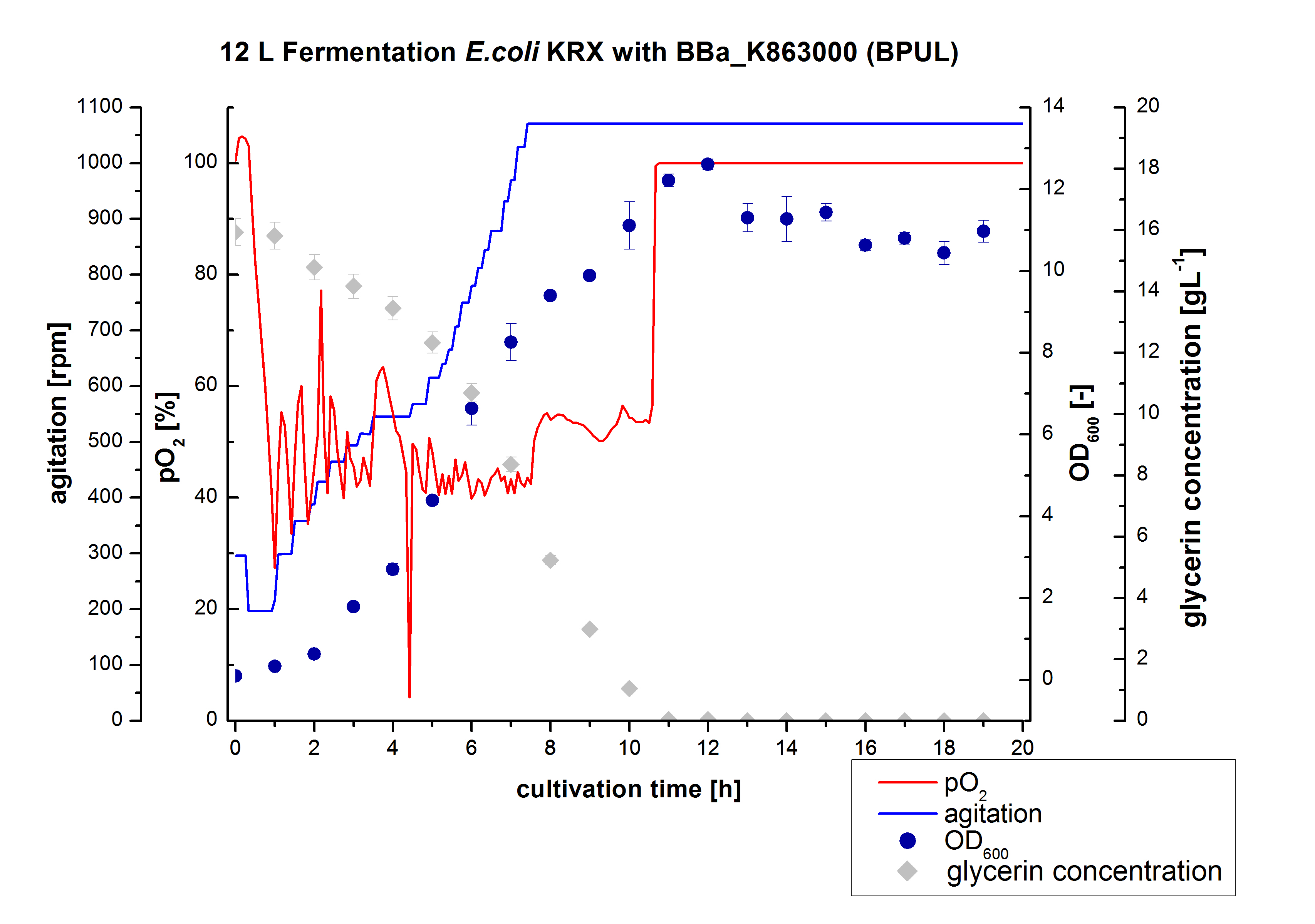
** 12L Cutlivations of ''E. coli'' KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] were harvested and stored at 4°C until purification. | ** 12L Cutlivations of ''E. coli'' KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] were harvested and stored at 4°C until purification. | ||
| + | |||
| + | [[File:Bielefeld2012_ECOL_Fermentation_12L.jpg|350px|thumb|left|'''Figure 1:''' Fermentation of ''E. coli'' KRX with <partinfo>BBa_K863005</partinfo> (ECOL) in an Bioengineering NLF 19, scale: 12 L, [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] + 60 µg/mL chloramphenicol, 37 °C, pH 7, agitation on cascade to hold pO<sub>2</sub> at 50 %, OD<sub>600</sub> measured every 30 minutes.]] | ||
| + | [[File:Bielefeld2012_BPUL_Fermentation_12L.jpg|350px|thumb|right|'''Figure 2:''' Fermentation of ''E. coli'' KRX with <partinfo>BBa_K863000</partinfo> (BPUL) in an Bioengineering NLF 19, scale: 12 L, [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] + 60 µg/mL chloramphenicol, 37 °C, pH 7, agitation on cascade to hold pO<sub>2</sub> at 50 %, OD<sub>600</sub> measured every 30 minutes.]] | ||
| + | <br style="clear: both" /> | ||
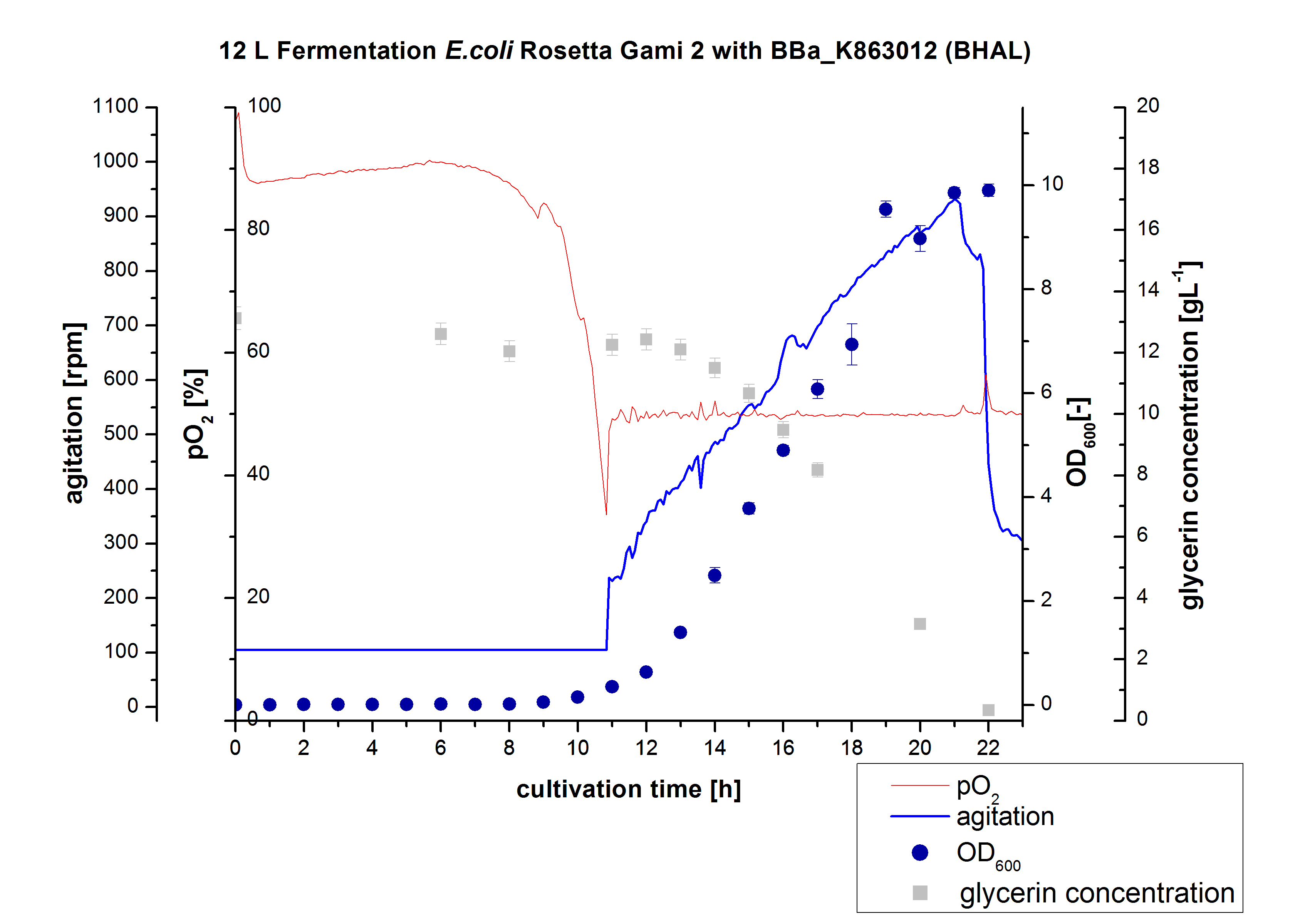
** 12 L Cultivation of ''E. coli'' Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) | ** 12 L Cultivation of ''E. coli'' Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) | ||
*** Settings: | *** Settings: | ||
Bioengineering NFL 19L fermenter, HSG medium, 60 µg/mL chloramphenicol, 300 µg/mL ampicillin, 12 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 12 NL/m, 22-24 hours. HSG medium was chosen to get a high biomass concentration with hope for a higher amount of laccases. | Bioengineering NFL 19L fermenter, HSG medium, 60 µg/mL chloramphenicol, 300 µg/mL ampicillin, 12 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 12 NL/m, 22-24 hours. HSG medium was chosen to get a high biomass concentration with hope for a higher amount of laccases. | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Dissolved estradiol, estrone and ethinyl estradiol in 50 % (v/v) acetonitrile with a concentration of 200 µg mL<sup>-1</sup> | ||
| + | ** Creating and measuring of negative controls for estradiol, estrone and ethinyl estradiol. Experimental setup: 2 µg substrate, 110 µL BR-buffer in 200 µL reaction volume. Measured at t<sub>0</sub> and after 3 hours. | ||
===Saturday October 13th=== | ===Saturday October 13th=== | ||
* '''Team Shuttle Vector:''' | * '''Team Shuttle Vector:''' | ||
| - | ** The P. pastoris GS115 cells integrated with | + | ** The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were also inducted with 0.5% (v/v) methanol (100%). |
* '''Team Cultivation and Purification''' | * '''Team Cultivation and Purification''' | ||
| - | ** 12L Cutlivations of ''E. coli'' Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) were harvested and stored at 4°C until purification. | + | ** 12L Cutlivations of ''E. coli'' Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) were harvested and stored at 4°C until purification. |
| + | |||
| + | [[File:Bielefeld2012 BHAL12L.jpg|350px|thumb|left|'''Figure 1:''' Fermentation of ''E. coli'' KRX with <partinfo>BBa_K863005</partinfo> (ECOL) in an Bioengineering NLF 19, scale: 12 L, [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] + 60 µg/mL chloramphenicol, 37 °C, pH 7, agitation on cascade to hold pO<sub>2</sub> at 50 %, OD<sub>600</sub> measured every 30 minutes.]] | ||
| + | [[File:Bielefeld2012 TTHL12L.jpg|350px|thumb|right|'''Figure 2:''' Fermentation of ''E. coli'' KRX with <partinfo>BBa_K863000</partinfo> (BPUL) in an Bioengineering NLF 19, scale: 12 L, [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] + 60 µg/mL chloramphenicol, 37 °C, pH 7, agitation on cascade to hold pO<sub>2</sub> at 50 %, OD<sub>600</sub> measured every 30 minutes.]] | ||
| + | <br style="clear: both" /> | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Creating and measuring of negative controls for estradiol, estrone and ethinyl estradiol. Experimental setup: 2 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer in 200 µL reaction volume. Measured at t<sub>0</sub> and after 10 minutes. | ||
| + | |||
| + | <br style="clear: both" /> | ||
===Sunday October 14th=== | ===Sunday October 14th=== | ||
* '''Team Shuttle Vector''': | * '''Team Shuttle Vector''': | ||
| - | ** The P. pastoris GS115 cells integrated with | + | ** The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were also inducted with 0.5% (v/v) methanol (100%). |
* '''Team Fungal and Plant Laccases:''' | * '''Team Fungal and Plant Laccases:''' | ||
| - | ** Plasmid isolation of pSB1C3:: | + | ** Plasmid isolation of pSB1C3::BBa_K863207 for transformation into P. pastoris GS115 was done. After isolation the plasmid was digested with [[Team:Bielefeld-Germany/Protocols/Materials#Used_enzymes |NotI]], purified with the PCR clean up kit and transformed via [[Team:Bielefeld-Germany/Protocols/molecular_genetics#Transformation_of_yeast_cells |protocol]] in P. pastoris GS115. |
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
| Line 183: | Line 200: | ||
</td></tr></table> | </td></tr></table> | ||
</html> | </html> | ||
| - | |||
===Monday October 15th=== | ===Monday October 15th=== | ||
* '''Team Shuttle Vector:''' | * '''Team Shuttle Vector:''' | ||
** There are no colonies of pBS1C3::BBa_K863202 KRX transformantion. | ** There are no colonies of pBS1C3::BBa_K863202 KRX transformantion. | ||
| - | ** The P. pastoris GS115 cells integrated with | + | ** The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were harvested and the supernatant was examined on GFP by fluorescence. But there was no GFP. |
| + | |||
* '''Team Fungal and Plant Laccases:''' | * '''Team Fungal and Plant Laccases:''' | ||
| - | ** Yeah, the yeast cells are growing. | + | ** Yeah, the yeast cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863207 BBa_K863207] are growing. |
| + | |||
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
** Tried to isolate the eight plated, but did get low concentrations again (except one J61101+GFP_Freiburg+CBDcex_Freiburg with conc >100 ng/µL) | ** Tried to isolate the eight plated, but did get low concentrations again (except one J61101+GFP_Freiburg+CBDcex_Freiburg with conc >100 ng/µL) | ||
** cleaned up some more PCR-products (GFP_Freiburg; CBDcex_Freiburg; CBDclos_Freiburg) | ** cleaned up some more PCR-products (GFP_Freiburg; CBDcex_Freiburg; CBDclos_Freiburg) | ||
*'''Team Substrate Analysis''': We stopped the reaction from the 12th October with Methanol and gave it to Dr. Marcus Persicke to measure the degradation on LC-MS | *'''Team Substrate Analysis''': We stopped the reaction from the 12th October with Methanol and gave it to Dr. Marcus Persicke to measure the degradation on LC-MS | ||
| + | |||
| + | * '''Team Cultivation and Purification''' | ||
| + | ** Harvested Cells from 12L Cutlivations of ''E. coli'' KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] as well as 12 L Cultivation of ''E. coli'' Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) were disrupted via high-pressure homogenizer and filtrated by [http://www.millipore.com/catalogue/module/C7493 Millipore Pellicon XL 50] with first 300 kDa to seperate all celldebris and 10 kDa to concentrate the laccases. The concentrated solutions were purified by the Ni-NTA-column. | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Creating and measuring of estradiol and ethinyl estradiol degradation samples for the laccase TVEL0. Experimental setup: 2 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 10 µg laccase in 200 µL reaction volume. Measured at t<sub>0</sub> and after 10 minutes. | ||
| + | |||
| + | * '''Team Immobilization''' | ||
| + | **Started immobilization of TVEL0 to analyze the binding of laccase during the first 10 hours representative for following immobilizations. | ||
===Tuesday October 16th=== | ===Tuesday October 16th=== | ||
| Line 207: | Line 233: | ||
** Plated both on AMP-select-agar | ** Plated both on AMP-select-agar | ||
** Plated one more Colony of J61101 + GFP_Freiburg and J61101 + GFP_Freiburg + CBDclos | ** Plated one more Colony of J61101 + GFP_Freiburg and J61101 + GFP_Freiburg + CBDclos | ||
| - | + | ||
| - | + | ||
| - | + | ||
*'''Team Substrate Analysis:''' The Massspectrometry data showed two potential products after the Laccase treatment of Estradiol and Ethinyl estradiol but we unfortunatly could not identify them so we decided to do MS-MS on thoose. The products showed that after Laccase treatment both Estradiol and Ethinyl estradiol losses two '''H''' atoms which was new for us. | *'''Team Substrate Analysis:''' The Massspectrometry data showed two potential products after the Laccase treatment of Estradiol and Ethinyl estradiol but we unfortunatly could not identify them so we decided to do MS-MS on thoose. The products showed that after Laccase treatment both Estradiol and Ethinyl estradiol losses two '''H''' atoms which was new for us. | ||
| + | |||
| + | *'''Team Immobilization:''' | ||
| + | Taking samples for immobilization time analyzation. | ||
===Wednesday October 17th=== | ===Wednesday October 17th=== | ||
| Line 216: | Line 243: | ||
** Before starting our activity assay we re-buffered the fractions we got from Team Cultivation and Purification. It took some time, that's why we had no time for activity analysis yet. But we prepared everything for the next day, including determining the protein concentration of the fractions after they have been re-buffered. This is what we got: | ** Before starting our activity assay we re-buffered the fractions we got from Team Cultivation and Purification. It took some time, that's why we had no time for activity analysis yet. But we prepared everything for the next day, including determining the protein concentration of the fractions after they have been re-buffered. This is what we got: | ||
[[File:Bielefeld2012_Prot_nach_Umpuffern.jpg|300px|thumb|center]] | [[File:Bielefeld2012_Prot_nach_Umpuffern.jpg|300px|thumb|center]] | ||
| + | |||
* '''Team Shuttle Vector:''' | * '''Team Shuttle Vector:''' | ||
| - | ** Genomic DNA isolation of yeast cells with integrated | + | ** Genomic DNA isolation of yeast cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] construct was done with the Kit. For phenotype characterisation an PCR with the primer pair 5AOX-Phenotype-FW and TT-Phenotype-RV was done and the fragment size was determined. The results are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] was not integrated. |
| + | |||
* '''Team Fungal and Plant Laccases:''' | * '''Team Fungal and Plant Laccases:''' | ||
| - | ** YPD liquid culture was inoculated with one yeast colony ( | + | ** YPD liquid culture was inoculated with one yeast colony ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863207 BBa_K863207]) and incubated at 30°C and 150 rpm. |
| + | |||
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
** Picked colonies of B0034 and J23100 and plated them on AMP selection agar for plasmid isolation | ** Picked colonies of B0034 and J23100 and plated them on AMP selection agar for plasmid isolation | ||
| + | |||
* '''Team Site Directed Mutagenesis:''' | * '''Team Site Directed Mutagenesis:''' | ||
** SDM-PCR of the Shuttle vector | ** SDM-PCR of the Shuttle vector | ||
** Added ''Dpn''I over night | ** Added ''Dpn''I over night | ||
| + | |||
| + | * '''Team Cultivation and Purification:''' | ||
| + | ** Made SDS-Pages of the cultivations from 10/11 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]) and 10/12 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012], <partinfo>BBa_K863022</partinfo>). | ||
| + | [[File:Bielefeld2012_1019.jpg|600px|thumb|center|'''Figure 1:''' SDS-Page of purification from the 12 L fermentations from 10/11 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]) and 10/12 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012], <partinfo>BBa_K863022</partinfo>). Purification of the supernatant via His-trap column, step gradient with 5 %, 50 % and 100 % elution buffer.]] | ||
| + | |||
| + | * '''Team Immobilization:''' | ||
| + | Measurement of Protein concentration with [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Used_Kits Roti-Nanoquant] and analyses of binding capacity. | ||
===Thursday October 18th=== | ===Thursday October 18th=== | ||
* '''Team Activity Tests:''' | * '''Team Activity Tests:''' | ||
** Finally the day had come to measure the activity of our own produced laccases. This activity assay should give information about the enzyme content in every fraction. To make the measurements comparable we applied the same protein amount in every sample. Again, we used our [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol], but this time we used the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Britton-Robinson_Buffer/ Britton-Robinson Buffer] at pH 5. Also we applied 0.1 mM ABTS and measured over night. | ** Finally the day had come to measure the activity of our own produced laccases. This activity assay should give information about the enzyme content in every fraction. To make the measurements comparable we applied the same protein amount in every sample. Again, we used our [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol], but this time we used the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Britton-Robinson_Buffer/ Britton-Robinson Buffer] at pH 5. Also we applied 0.1 mM ABTS and measured over night. | ||
| - | [[File:Bielefeld2012_new_ECOL_activity.jpg|360px|thumb|left|Activity assay of each purified fraction of our new cultivation with ECOL. Samples were re-buffered into H<sub>2</sub> and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]][[File:Bielefeld2012_new_BPUL_acitivity.jpg|360px|thumb|right|Activity assay of each purified fraction of our new cultivation with BPUL. Samples were re-buffered into H<sub>2</sub> and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]] | + | [[File:Bielefeld2012_new_ECOL_activity.jpg|360px|thumb|left|Activity assay of each purified fraction of our new cultivation with ECOL. Samples were re-buffered into H<sub>2</sub>O and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]][[File:Bielefeld2012_new_BPUL_acitivity.jpg|360px|thumb|right|Activity assay of each purified fraction of our new cultivation with BPUL. Samples were re-buffered into H<sub>2</sub>O and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]] |
| - | [[File:Bielefeld2012_new_BHAL_activity.jpg|360px|thumb|left|Activity assay of each purified fraction of our new cultivation with BHAL. Samples were re-buffered into H<sub>2</sub> and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]][[File:Bielefeld2012_new_TTHL_activity.jpg|360px|thumb|right|Activity assay of each purified fraction of our new cultivation with TTHL. Samples were re-buffered into H<sub>2</sub> and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]] | + | [[File:Bielefeld2012_new_BHAL_activity.jpg|360px|thumb|left|Activity assay of each purified fraction of our new cultivation with BHAL. Samples were re-buffered into H<sub>2</sub>O and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]][[File:Bielefeld2012_new_TTHL_activity.jpg|360px|thumb|right|Activity assay of each purified fraction of our new cultivation with TTHL. Samples were re-buffered into H<sub>2</sub>O and the protein amount in each fraction has been adjusted. The Measurement was done using the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Analytics#General_setup_of_enzyme_activity_measurements/ standard activity assay protocol] over night.]] |
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| Line 254: | Line 297: | ||
** Clean-Up of 19C-PRC-product | ** Clean-Up of 19C-PRC-product | ||
** Transformation of the 19C-PRC-product in XL1 Blue | ** Transformation of the 19C-PRC-product in XL1 Blue | ||
| + | |||
| + | * '''Team Immobilization:''' | ||
| + | **Discussed new method of activity measurements for immobilized laccases on beads with less lacasse solution needed. Started first general tests with photometer. | ||
===Friday October 19th=== | ===Friday October 19th=== | ||
| Line 271: | Line 317: | ||
** Restriction of Ecol_Freiburg with ''Xba''I, ''Age''I and ''Dpn''I | ** Restriction of Ecol_Freiburg with ''Xba''I, ''Age''I and ''Dpn''I | ||
* '''Team Site Directed Mutagenesis:''' | * '''Team Site Directed Mutagenesis:''' | ||
| + | |||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Creating and measuring of estradiol and ethinyl estradiol degradation samples for the laccases TVEL0 and BPUL. The experimental setup changed. The laccase amount was adjusted to the highest possible amount of the lowest concentrated laccase. TTHL has a concentration of 4,03 µg mL<sup>-1</sup> and sets the maximal laccase amount to 0.6 µg per 200 µL reaction volume. To compensate these worse conditions, the substrate amount was lowered to 1 µg and the temperature was changed to 30°C. | ||
| + | ** Test of estradiol and ethinyl estradiol degradation done by TVEL0 and BPUL without ABTS. Experimental setup: 1 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 0.6 µg laccase in 200 µL reaction volume. Measured at t<sub>0</sub> and after 3 hours. | ||
===Saturday October 20th=== | ===Saturday October 20th=== | ||
*'''Team Activity Tests:''' | *'''Team Activity Tests:''' | ||
| - | ** [[File:Bielefeld2012_Klein_ABTS_neg_control.jpg|thumb|300px|right|Negativ control for substrate saturation analysis to investigate in the effect of CuCl<sub>2</sub> | + | ** [[File:Bielefeld2012_Klein_ABTS_neg_control.jpg|thumb|300px|right|Negativ control for substrate saturation analysis to investigate in the effect of CuCl<sub>2</sub> oxidizing ABTS. Instead of laccase in H<sub>2</sub>O, only H<sub>2</sub>O mixed with 0.4 mM CuCl<sub>2</sub> was applied.]] During the Jamboree in Amsterdam we had the oppurtunity to discuss our activity results with a lot of people. One remark was about the calculation of the enzyme specific units. As you all remember, we measured the activty with 0.1 mM ABTS, but to get comparable results with the literature, we had to measure each protein with other concentrations of ABTS. The concentration of ABTS should be saturated for each laccase. So we started with ECOL and BPUL and applied different ABTS concentrations starting from 0.1 mM as usual: 0.1 mM, 0.5 mM, 1 mM and 5 mM. We applied 616 ng Laccase in each sample. Our results showed, that especially ECOL does not reach its substrate saturation with 5 mM ABTS. Our plans are now to check these concentrations of ABTS for BPUL and TTHL and also higher concentrations of ABTS for all laccases. For that we should reduce the enzyme amount to be able to detect the change in OD 420 properly. |
| - | [[File:Bielefeld2012_ECOL_klein_ABTS.jpg|thumb|360px|right|Activity assay of our new ECOL laccase with different ABTS concentration to find the substrate saturation of ECOL. 616 ng ECOL laccase (incubated with 0.4 mM CuCl<sub>2</sub>) and 20 % Britton-Robinson buffer(pH 5) were applied.]][[File:Bielefeld2012_BPUL_klein_ABTS.jpg|thumb|360px|left|Activity assay of our new BPUL laccase with different ABTS concentration to find the substrate saturation of BPUL. 616 ng BPUL laccase (incubated with 0.4 mM CuCl<sub>2</sub> and 20 % Britton-Robinson buffer(pH 5) were applied.]]<br>< | + | [[File:Bielefeld2012_ECOL_klein_ABTS.jpg|thumb|360px|right|Activity assay of our new ECOL laccase with different ABTS concentration to find the substrate saturation of ECOL. 616 ng ECOL laccase (incubated with 0.4 mM CuCl<sub>2</sub>) and 20 % Britton-Robinson buffer (pH 5) were applied.]][[File:Bielefeld2012_BPUL_klein_ABTS.jpg|thumb|360px|left|Activity assay of our new BPUL laccase with different ABTS concentration to find the substrate saturation of BPUL. 616 ng BPUL laccase (incubated with 0.4 mM CuCl<sub>2</sub>) and 20 % Britton-Robinson buffer (pH 5) were applied.]] |
| + | <br style="clear: both" /> | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** TVEL0 and BPUL were able to degrade 100 % of estradiol and 100 % of ethinyl estradiol. To get comparable results, the reaction temperature was lowered to 20 °C again, to have a closer look, where differences are apparent. Again TVEL0 and BPUL degraded 100 % of the substrates. But before the laccase stocks deplete, first ECOL, BHAL and TTHL should be tested. | ||
| + | ** Did substrate degradation analysis with ECOL without ABTS. Experimental setup: 1 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 0.6 µg laccase in 200 µL reaction volume. Measured at t<sub>-0</sub> and after 3 hours. | ||
===Sunday October 21st=== | ===Sunday October 21st=== | ||
| + | * '''Team Fungal and Plant Laccases:''' | ||
| + | Genomic isolation was done with the [http://www.promega.com/resources/protocols/technical-manuals/0/wizard-genomic-dna-purification-kit-protocol/ Promega Wizard genomic DNA purification system kit] of our approximately 50 yeast clones with integrated (a) GFP and (b) tvel5. | ||
* '''Team Activity Tests:''' | * '''Team Activity Tests:''' | ||
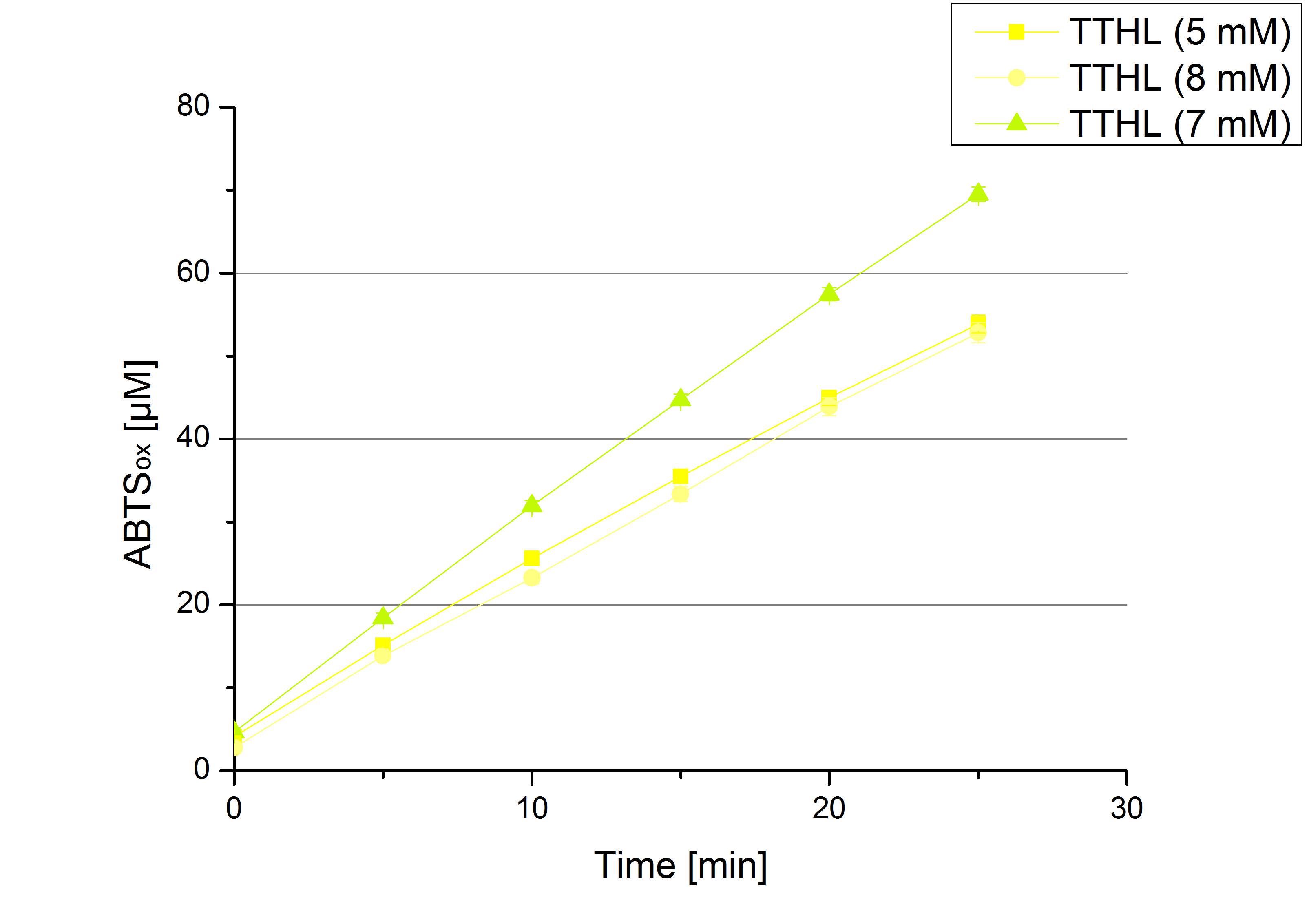
| - | ** Today was BHAL's and TTHL's turn. The experiment setup was the same as described the day before. But again we could not make sure, that 5 mM | + | ** Today was BHAL's and TTHL's turn. The experiment setup was the same as described the day before. But again we could not make sure, that 5 mM ABTS is the substrate saturation for our tested laccases. We are now sure to go higher in ABTS concentrations. |
| - | [[File:Bielefeld2012_BHAL_klein_ABTS.jpg|thumb|left|360px|Activity assay of our new BHAL laccase with different ABTS concentration to find the substrate saturation of BHAL. 616 ng ECOL laccase (incubated with 0.4 mM CuCl<sub>2</sub> and 20 % Britton-Robinson buffer | + | [[File:Bielefeld2012_BHAL_klein_ABTS.jpg|thumb|left|360px|Activity assay of our new BHAL laccase with different ABTS concentration to find the substrate saturation of BHAL. 616 ng ECOL laccase (incubated with 0.4 mM CuCl<sub>2</sub> and 20 % Britton-Robinson buffer pH 5) were applied.]][[File:Bielefeld2012_TTHL_klein_ABTS.jpg|thumb|right|360px|Activity assay of our new TTHL laccase with different ABTS concentration to find the substrate saturation of TTHL. 616 ng TTHL laccase (incubated with 0.4 mM CuCl<sub>2</sub> and 20 % Britton-Robinson buffer pH 5) were applied.]]<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> |
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
| Line 293: | Line 349: | ||
** Isolated the plasmids of the six SDM-dishes | ** Isolated the plasmids of the six SDM-dishes | ||
** Over-night digestion with ''Not''I | ** Over-night digestion with ''Not''I | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** ECOL, BHAL and TTHL were tested with the same conditions and all of them degraded the substrates much slower than TVEL0 and BPUL. | ||
| + | *''' Team Immobilization''' | ||
| + | **Started immobilization of new laccases ECOL, BPUL, BHAL, TTHL and TVEL0. | ||
| Line 326: | Line 386: | ||
</td></tr></table> | </td></tr></table> | ||
</html> | </html> | ||
| - | |||
===Monday October 22nd=== | ===Monday October 22nd=== | ||
| + | * '''Team Fungal and Plant Laccases:''' | ||
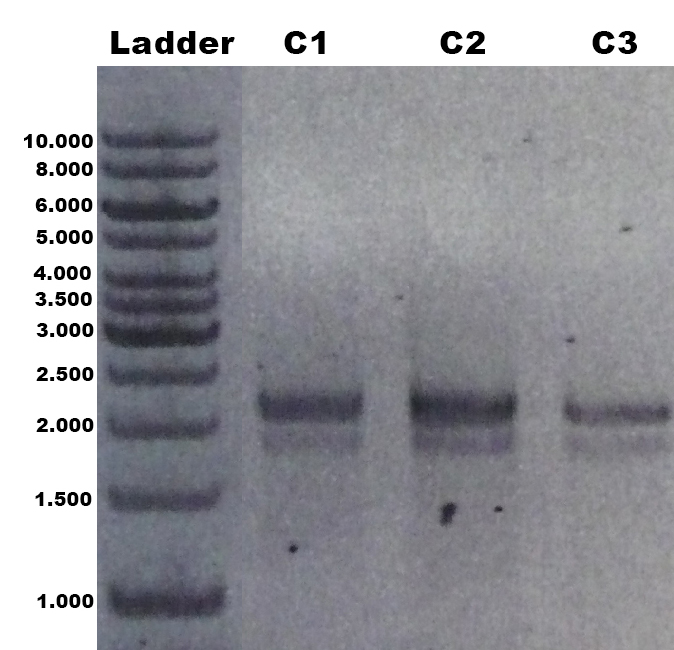
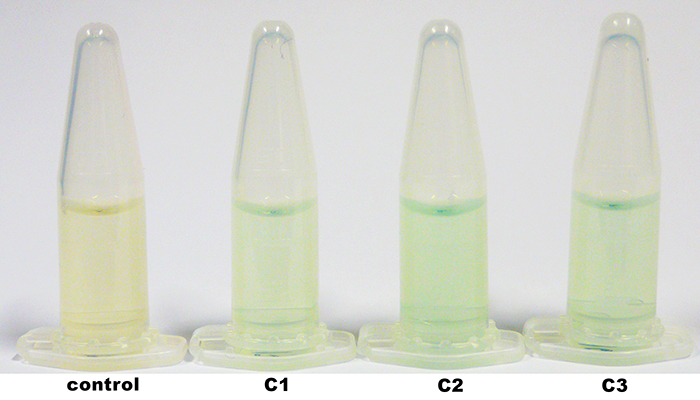
| + | The identification of positive yeast clones and determination of the phenotype (M<sup>+</sup> or M<sup>S</sup>) was done by PCR with the primers 5AOX-Phenotype-FW and TT-Phenotype-RV. The result of the PCR was: no GFP integrated clones and three tvel5 integrated (1.9 kb and 2.2 kb) clones. The cassette, including ''tvel5'' and ''his4'', was recombinated by a single cross over, because the 2.2 kb PCR product of the ''aox1'' gene has been formed. [[Team:Bielefeld-Germany/Protocols/Materials#Minimal_methanol_media_.28MM.29 | Minimal methanol medium]] was inoculated with these three positive clones with the M<sup>+</sup> phenotype. | ||
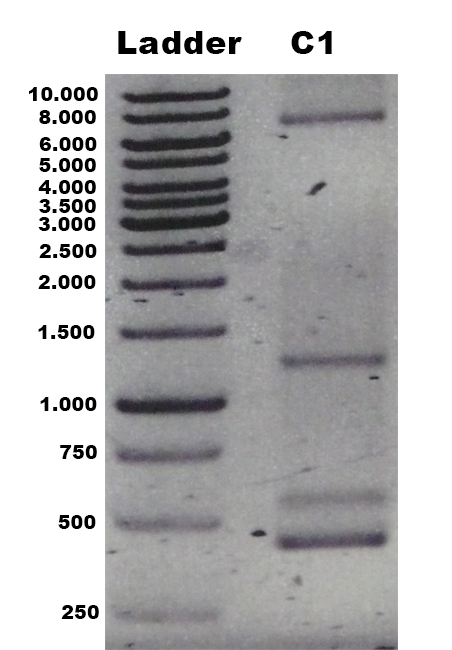
| + | [[File:Bielefeld2012_TV5_negKon_pSH19A_Agarosegel.jpg|thumb|left|150px|[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863204 BBa_K863204] was used as a controll for PCR with the primer 5AOX-Phenotype-FW and TT-Phenotype-RV. Expected lane: 437 bp.]] | ||
| + | [[File:Bielefeld2012_TV5_Probe_Agarosegel.jpg|thumb|right|250px|PCR product of the gDNA from the culture 1, 2 and 3 (C1, C2, C3) with the primer 5AOX-Phenotype-FW and TT-Phenotype-RV. Expected lane: 1.9 kb (''tvel5'') and 2.2 kb (''aox1'').]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
* '''Team Activity Tests:''' | * '''Team Activity Tests:''' | ||
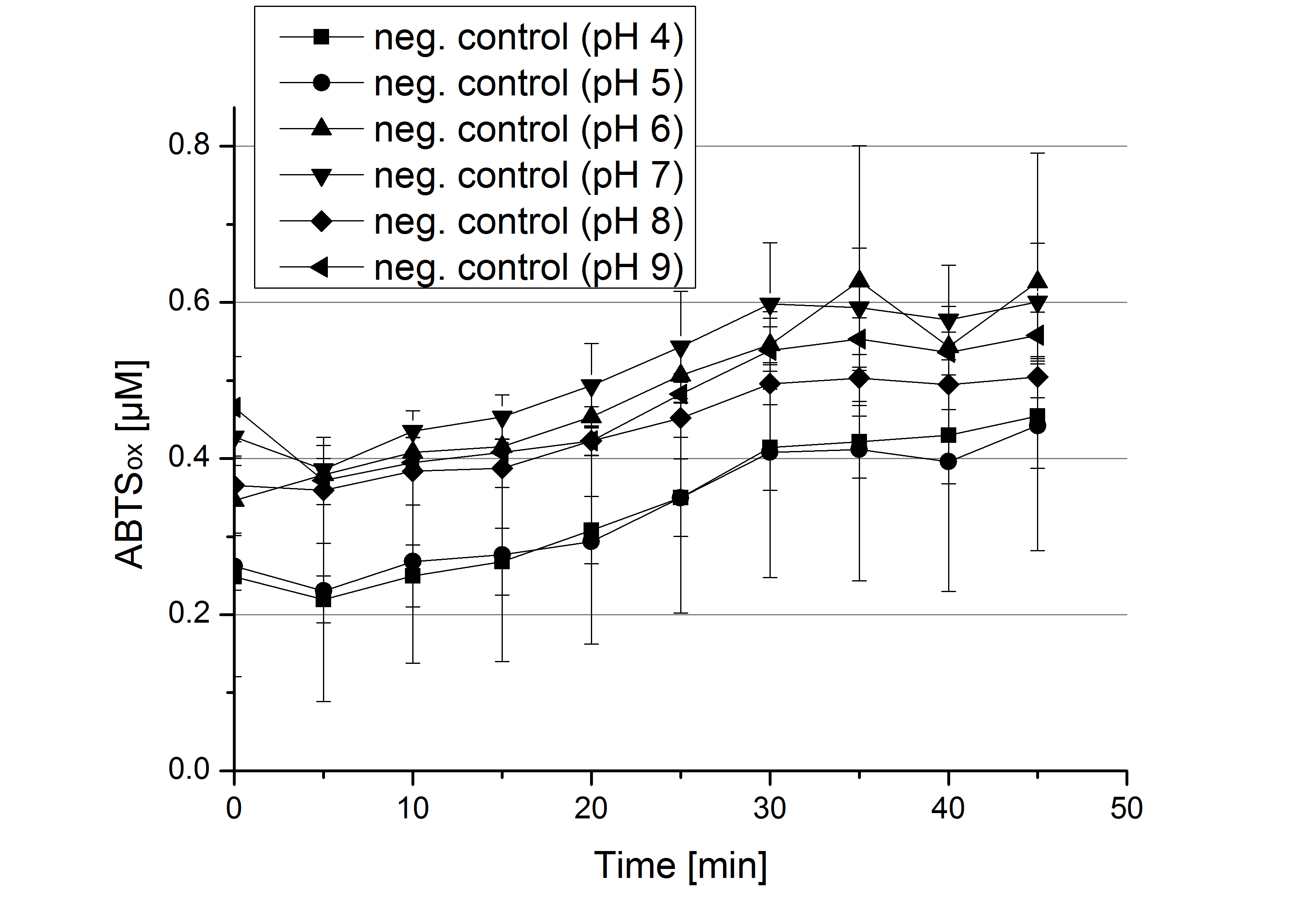
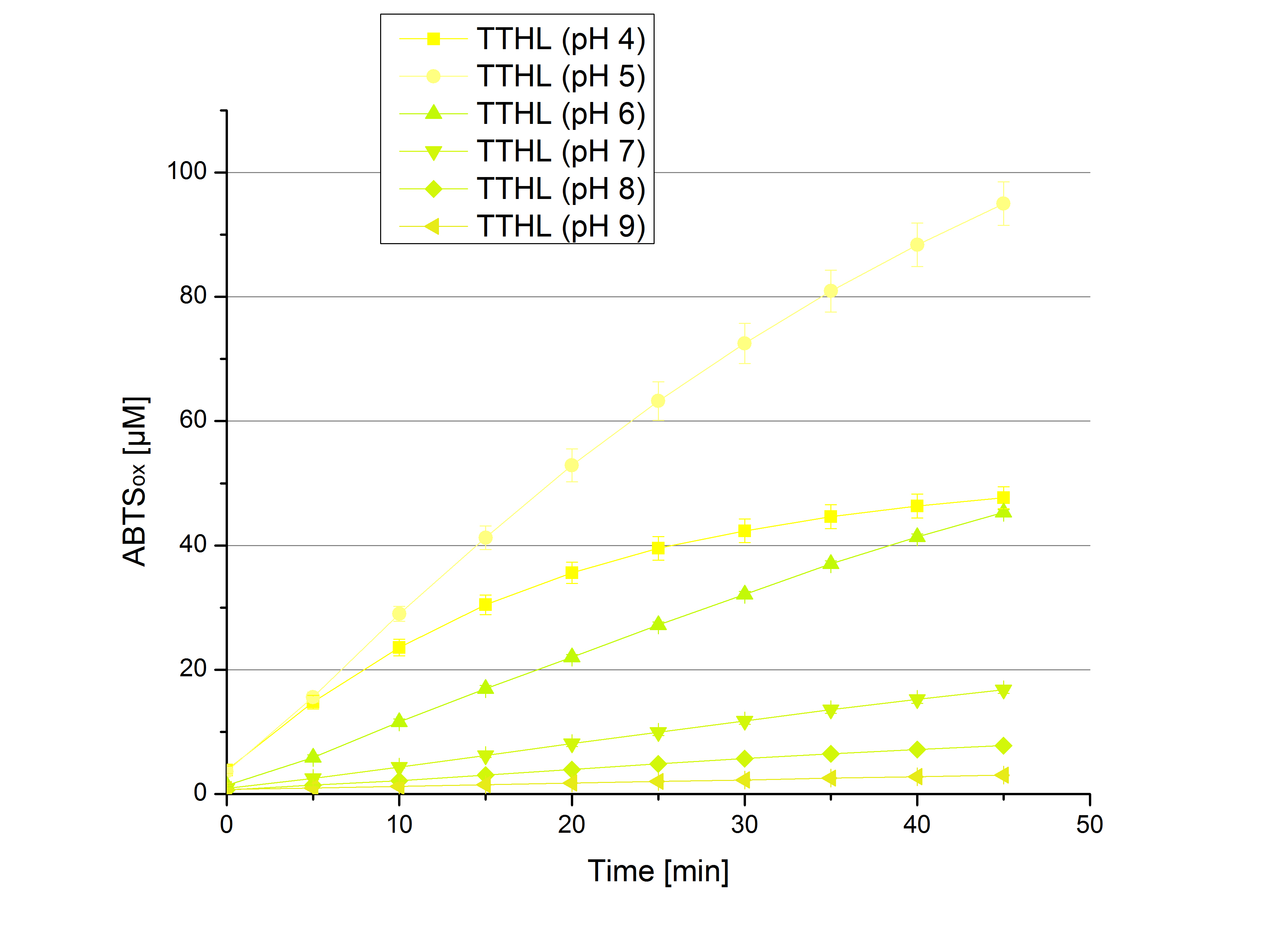
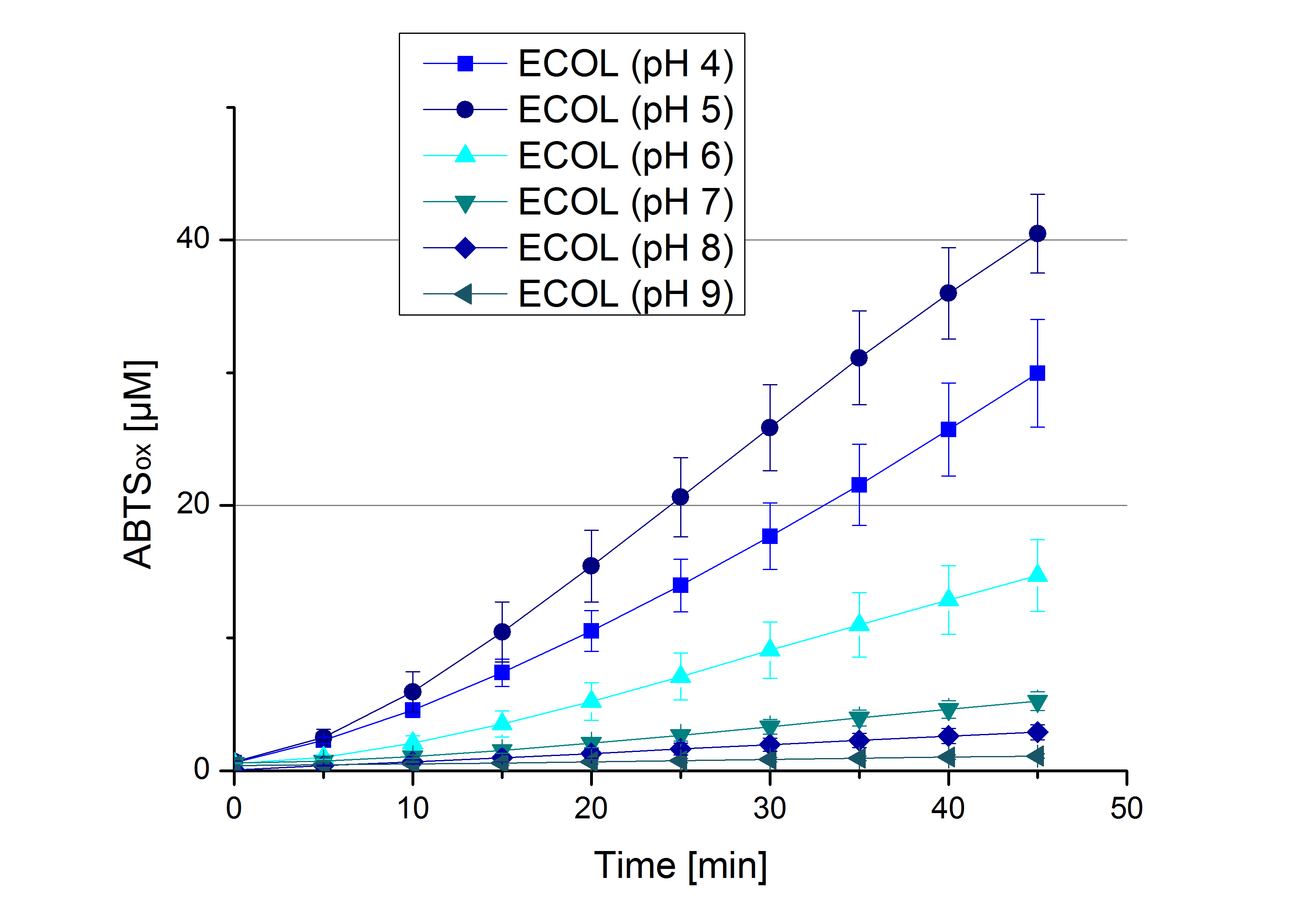
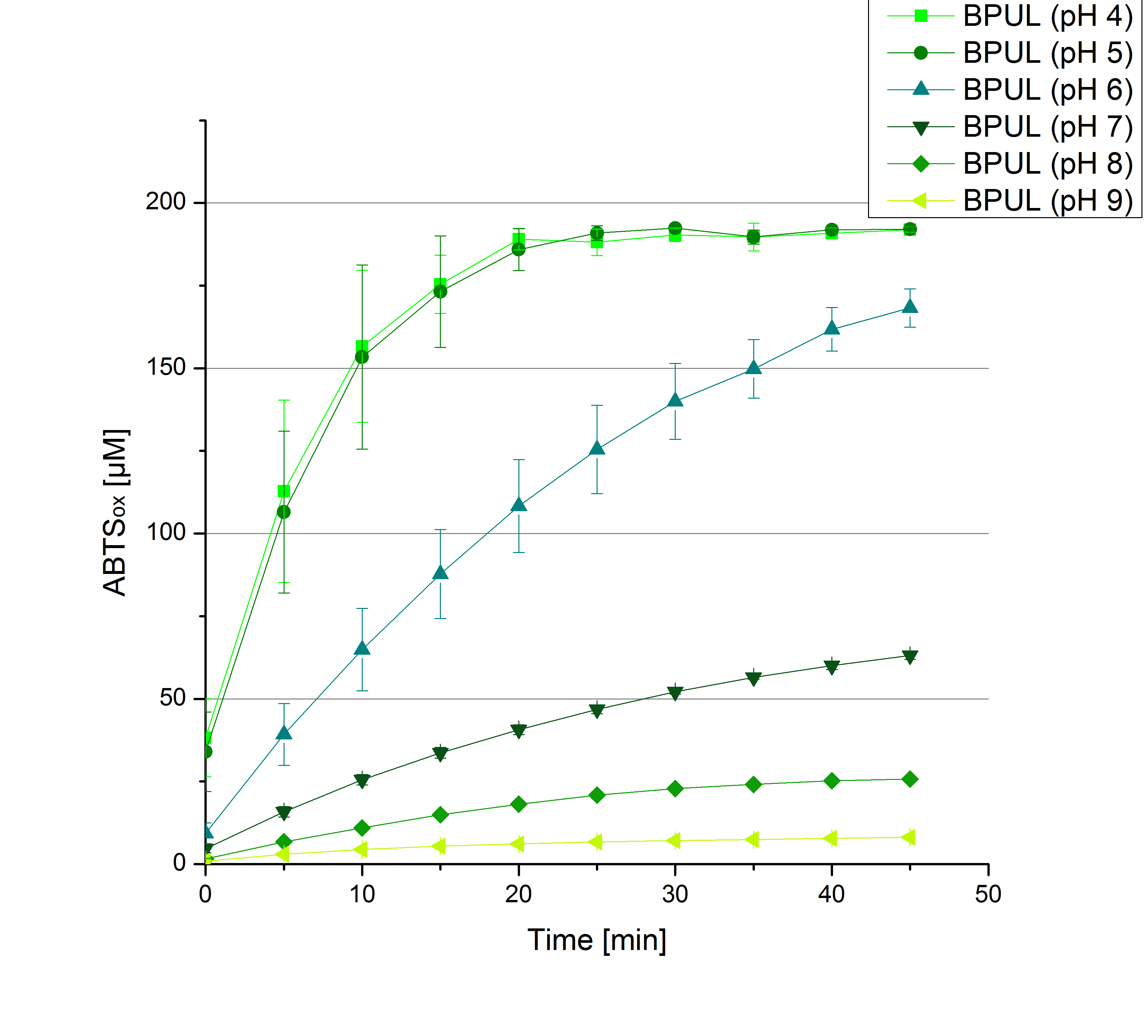
| - | ** [[File:Bielefeld2012_Neger_Kontrolle_pH_test.jpg|thumb|right|250px| | + | ** [[File:Bielefeld2012_Neger_Kontrolle_pH_test.jpg|thumb|right|250px|Negative control for the experimental setup with different pHs of the Britton-Robinson buffer. Effect of 5 mM ABTS was determined.]]Today was all about finding a comfy environment for our laccases. We thought of determining the right pH before going on with substrate saturation experiments since we could not be sure, that the activity was influenced by the pH. We chose a range from pH 4 to pH 9, applied 616 ng laccase and 5 mM ABTS. Our results revealed the following results: |
*** optimal pH of ECOL: pH 5 | *** optimal pH of ECOL: pH 5 | ||
*** optimal pH of BPUL: pH 4 and pH 5 | *** optimal pH of BPUL: pH 4 and pH 5 | ||
*** optimal pH of BHAL: pH 5 | *** optimal pH of BHAL: pH 5 | ||
*** optimal pH of TTHL: pH 5 | *** optimal pH of TTHL: pH 5 | ||
| - | <br><br><br> | + | <br><br><br><br> |
| - | [[File: | + | [[File:Bielefeld2012_BHAL_pH_test.jpg|thumb|left|360px|BHAL activity measured under different pH conditions (pH 4 to pH 9) to determine the pH optimum. 616 ng Laccase and 5 mM ABTS were used.]][[File:Bielefeld2012_TTHL_pH_test.jpg|thumb|right|360px|TTHL activity measured under different pH conditions (pH 4 to pH 9) to determine the pH optimum. 616 ng Laccase and 5 mM ABTS were used.]] |
| - | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | + | [[File:Bielefeld2012_ECOL_pH_test.jpg|thumb|left|360px|ECOL activity measured under different pH conditions (pH 4 to pH 9) to determine the pH optimum. 616 ng Laccase and 5 mM ABTS were used.]][[File:Bielefeld2012_BPUL_pH_test.jpg|thumb|right|360px|BPUL activity measured under different pH conditions (pH 4 to pH 9) to determine the pH optimum. 616 ng Laccase and 5 mM ABTS were used.]] |
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| Line 355: | Line 421: | ||
* '''Team Site Directed Mutagenesis:''' | * '''Team Site Directed Mutagenesis:''' | ||
** Stopped Digestion of the shuttle vector, but gel went wrong and the products were lost | ** Stopped Digestion of the shuttle vector, but gel went wrong and the products were lost | ||
| + | |||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** BHAL and TTHL were tested with the same conditions and all of them degraded the substrates much slower than TVEL0 and BPUL. | ||
| + | |||
| + | *''' Team Immobilization:''' | ||
| + | ** Measured of protein concentration in supernatants of immobilized ECOL, BPUL, BHAL, TTHL and TVEL0 with [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Used_Kits Roti-Nanoquant]. Analysed binding capacity of the different laccases. | ||
===Tuesday October 23rd=== | ===Tuesday October 23rd=== | ||
| + | * '''Team Fungal and Plant Laccases:''' The induction of the yeast cells with 1% (v/v) 100% methanol was done at morning and in the evening. | ||
| + | |||
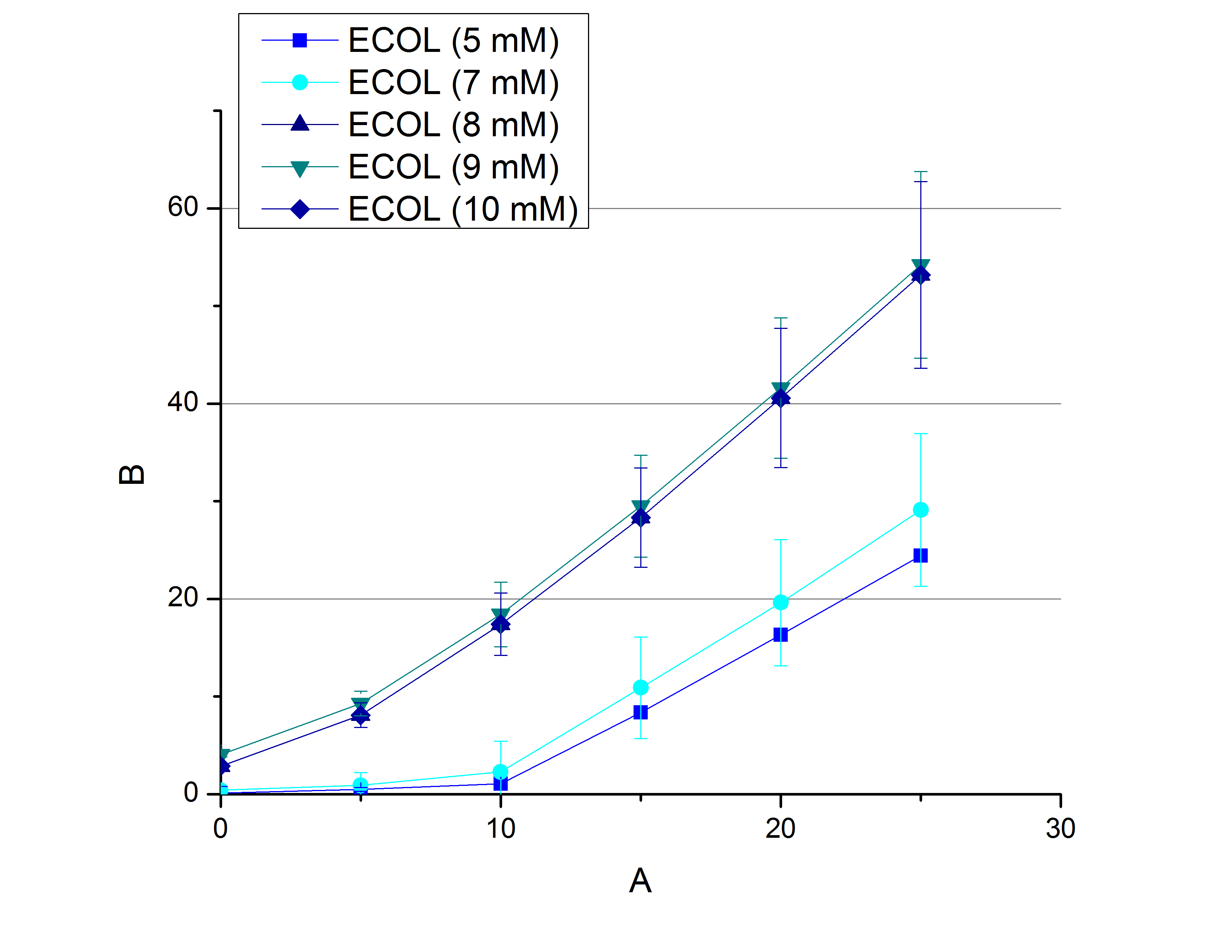
| + | * '''Team Activity Tests:''' | ||
| + | ** In order to find the substrate saturation we continued to measure our laccase activities with higher concentrations of ABTS. To have a slower reaction and also try not to waste the enzymes we halfed the amount of used laccases from 616 ng to 308 ng of each laccase. The following substrate saturations were determined: | ||
| + | *** ECOL: 9 mM ABTS | ||
| + | *** BPUL: 5 mM ABTS | ||
| + | *** BHAL: 7 mM ABTS | ||
| + | *** TTHL: 7 mM ABTS | ||
| + | |||
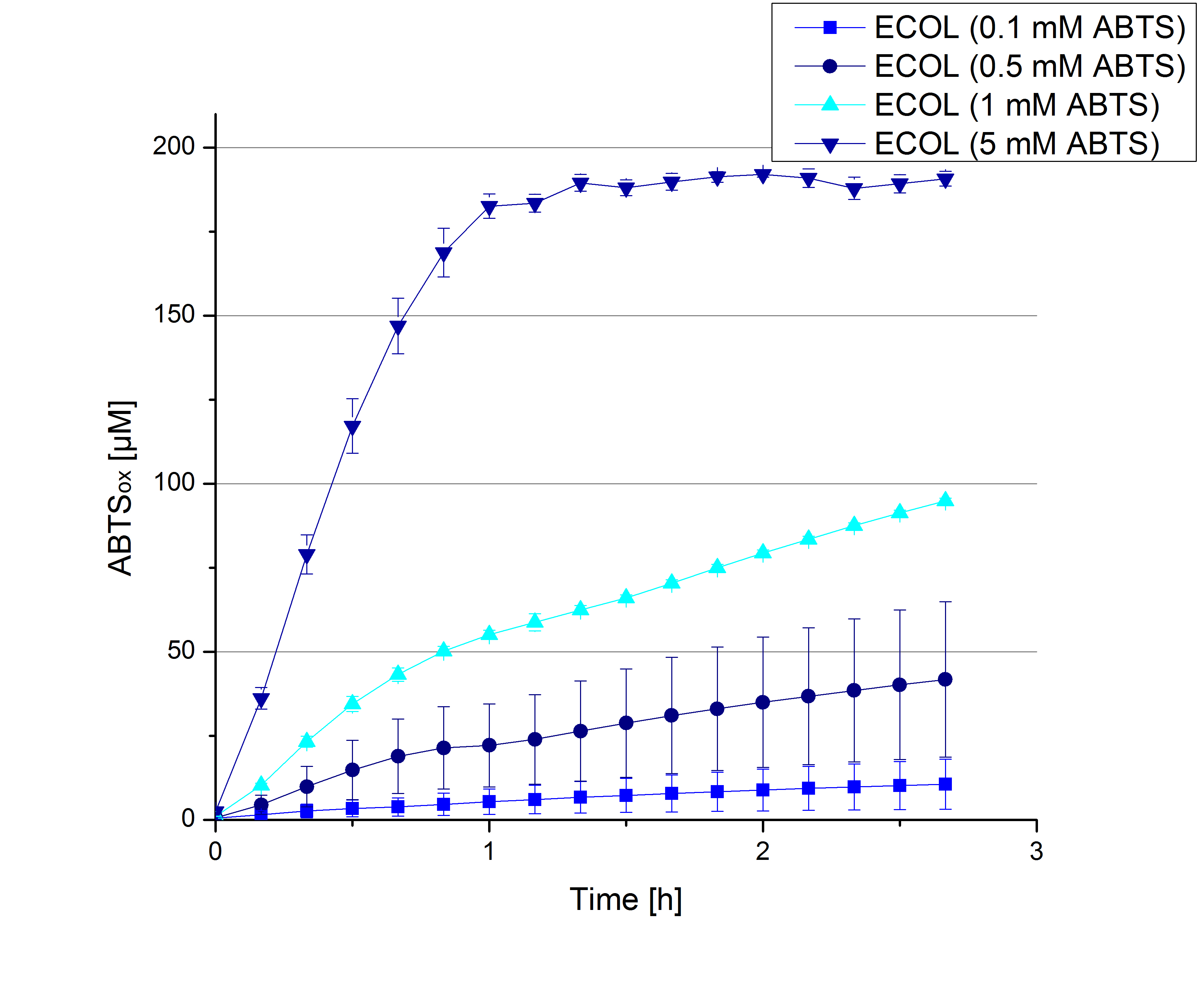
| + | [[File:Bielefeld2012_ECOL_hoch.jpg|thumb|left|360px]][[File:Bielefeld2012_BPUL_hoch.jpg|thumb|right|360px]] | ||
| + | <br style="clear: both" /> | ||
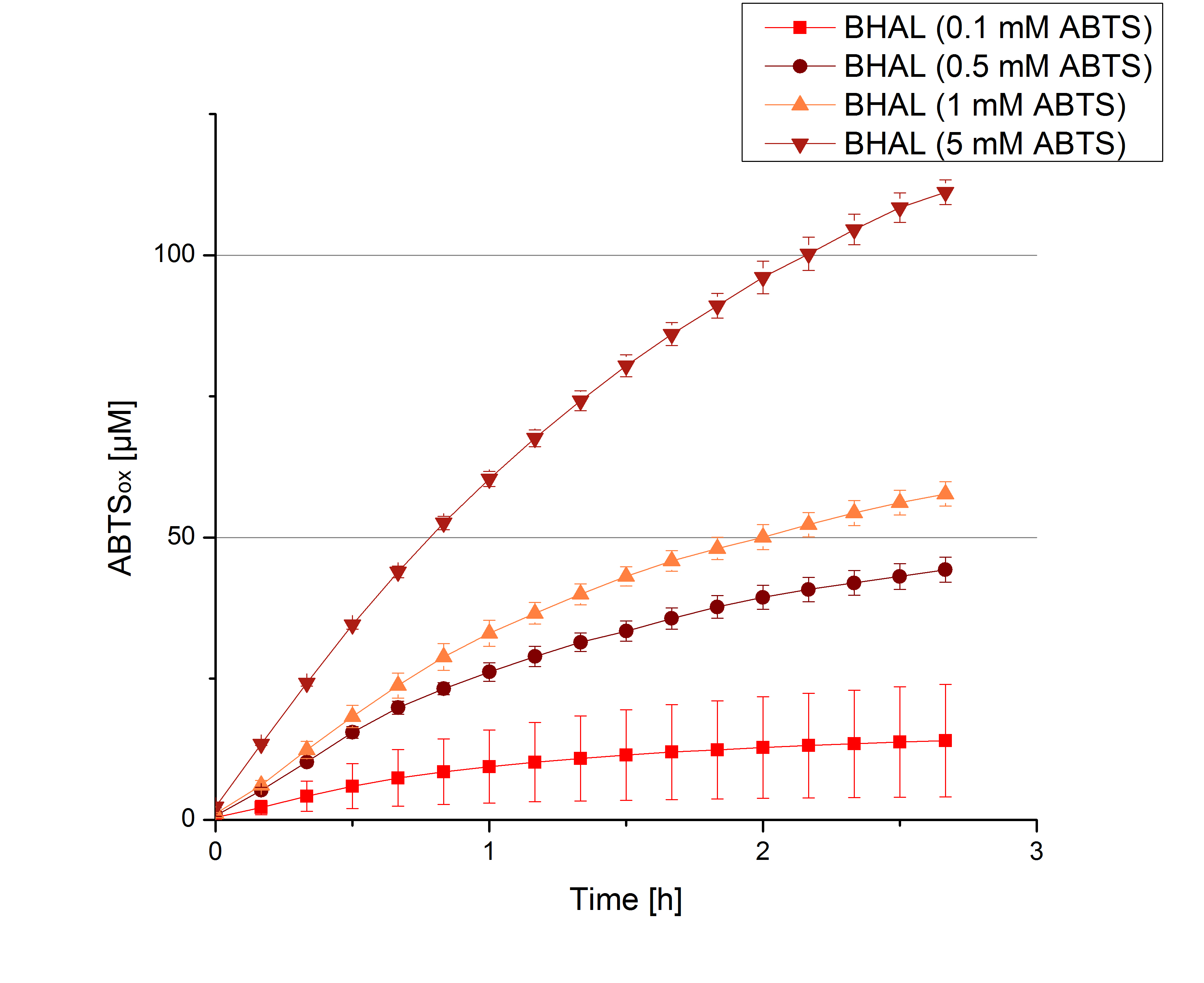
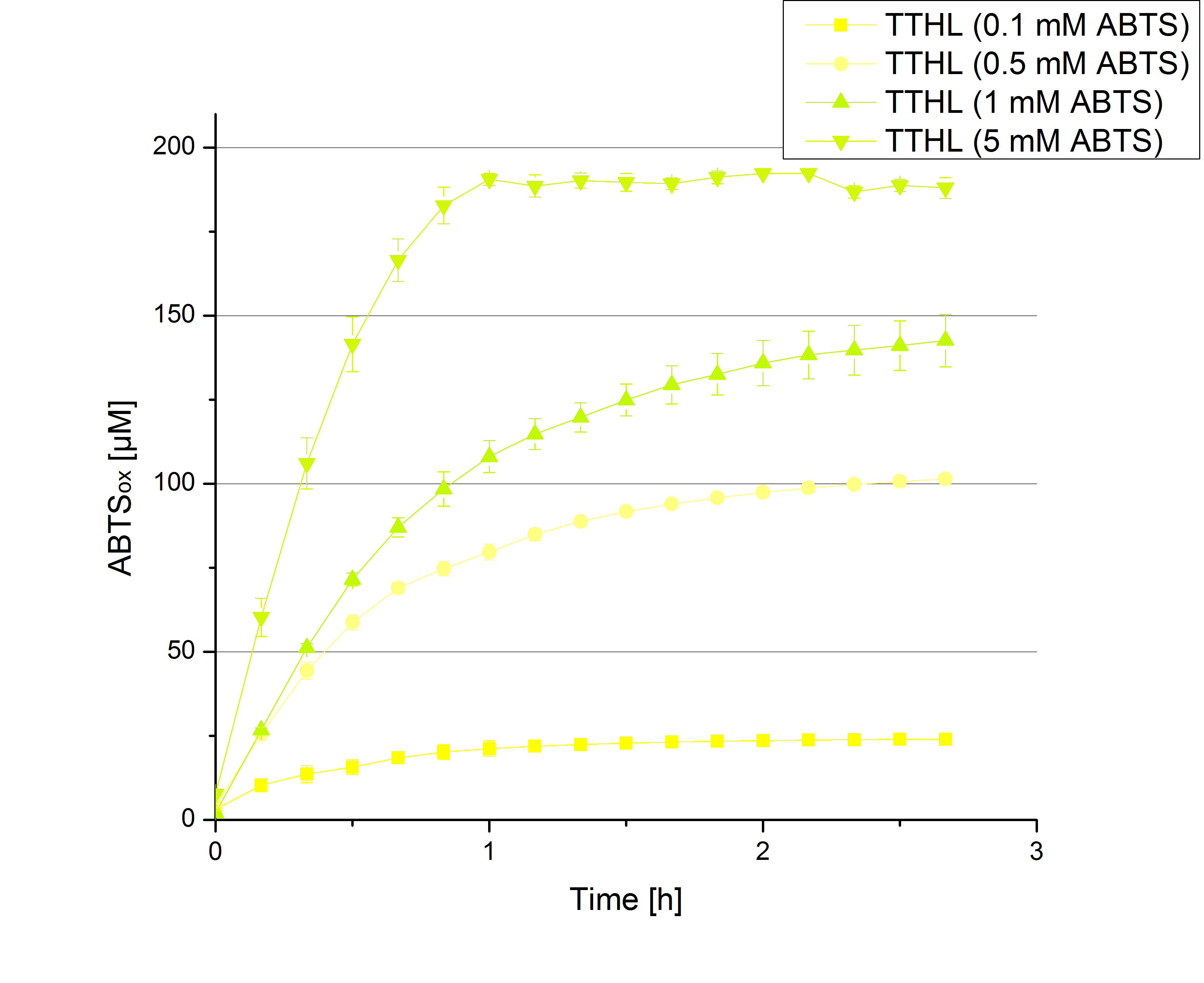
| + | [[File:Bielefeld2012_BHAL_ABTS_hoch.jpg|thumb|left|360px]][[File:Bielefeld2012_TTHL_hoch.jpg|thumb|right|360px]] | ||
| + | |||
| + | <br style="clear: both" /> | ||
| + | |||
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
** Colony PCR of B0034 + GFP_Freiburg | ** Colony PCR of B0034 + GFP_Freiburg | ||
| Line 364: | Line 451: | ||
*** 16/24 positive | *** 16/24 positive | ||
*** Plated three on selection agar for plasmid isolation | *** Plated three on selection agar for plasmid isolation | ||
| + | *'''Team Immobilization''' | ||
| + | **Started measurement of enzymatic activity of the different immobilized laccases with the [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Immobilization#Activity_measurement_of_beads Photometer]. | ||
===Wednesday October 24th=== | ===Wednesday October 24th=== | ||
| + | * '''Team Activity Tests:''' | ||
| + | ** pH measurements were repeated under substrate saturation with a pH range from pH 4 to pH 9. | ||
| + | |||
| + | * '''Team Fungal and Plant Laccases:''' In the morning and evening the induction of yeast cells with 1% (v/v) methanol was done. | ||
| + | |||
* '''Team Cellulose Binding Domain:''' | * '''Team Cellulose Binding Domain:''' | ||
** Isolated Plasmids containing B0034 + GFP_Freiburg & | ** Isolated Plasmids containing B0034 + GFP_Freiburg & | ||
| Line 374: | Line 468: | ||
** Clean up of inserts (B0034 + GFP_Freiburg & B0034 + CBDcex-GFP_Freiburg) and Backbone (J23100) via gel | ** Clean up of inserts (B0034 + GFP_Freiburg & B0034 + CBDcex-GFP_Freiburg) and Backbone (J23100) via gel | ||
** Ligation of Backbone and the inserts | ** Ligation of Backbone and the inserts | ||
| - | ** Transformation and | + | ** Transformation and plated on selection agar |
* '''Team Site Directed Mutagenesis:''' | * '''Team Site Directed Mutagenesis:''' | ||
** Digestion showed that all isolated colonies still have the illegal ''Xba''I restriction side | ** Digestion showed that all isolated colonies still have the illegal ''Xba''I restriction side | ||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Repeating of the degradation experiments without ABTS for TVEL0, BPUL, ECOL, BHAL and TTHL with a degradation time of five hours to increase the differences between the laccases. | ||
===Thursday October 25th=== | ===Thursday October 25th=== | ||
| - | *'''Team Fungal and Plant Laccases:''' Induction of the yeast cells with 1% (v/v) methanol was done. After 4 h the cells were harvested by centrifugation at 3000xg 4°C for 5 min | + | *'''Team Fungal and Plant Laccases:''' |
| + | ** Induction of the yeast cells with 1% (v/v) methanol was done. After 4 h the cells were harvested by centrifugation at 3000xg 4°C for 5 min. | ||
| - | *'''Team Substrate Anaylsis:''' The MS-MS data arrived from Marcus Persicke. To ensure our measurements and discuss what happens after the Laccase treatment and on the ESI we decided to ask an organic chemistrer so we went to Prof. Dr. Dietmar Kuck whos expert on this field. Since he had less time to discuss we have only speculativ testifies. | + | * '''Team Cellulose Binding Domain:''' |
| + | ** All colonies did not show any obvious green glow | ||
| + | |||
| + | *'''Team Substrate Anaylsis:''' | ||
| + | **The MS-MS data arrived from Marcus Persicke. To ensure our measurements and discuss what happens after the Laccase treatment and on the ESI we decided to ask an organic chemistrer so we went to Prof. Dr. Dietmar Kuck whos expert on this field. Since he had less time to discuss we have only speculativ testifies. | ||
| + | |||
| + | |||
| + | *'''Team Cultivation and Purification:''' | ||
| + | Last offline analysis of the the two cultivations of ''E.coli'' KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] with Carobon Source Detection with HPLC. | ||
| + | |||
| + | * '''Team Substrate Analysis:''' | ||
| + | ** Repeating of the degradation experiments with ABTS for ECOL, BHAL and TTHL to replace previous experimental errors. | ||
| + | |||
| + | ===Thursday October 25th=== | ||
| + | *'''Team Fungal and Plant Laccases:''' | ||
| + | ** Induction of the yeast cells with 1% (v/v) methanol was done. After 4 h the cells were harvested by centrifugation at 3000xg 4°C for 5 min. | ||
| + | |||
| + | * '''Team Cellulose Binding Domain:''' | ||
| + | ** All colonies did not show any obvious green glow | ||
| + | |||
| + | *'''Team Substrate Anaylsis:''' | ||
| + | **The MS-MS data arrived from Marcus Persicke. To ensure our measurements and discuss what happens after the Laccase treatment and on the ESI we decided to ask an organic chemistrer so we went to Prof. Dr. Dietmar Kuck whos expert on this field. Since he had less time to discuss we have only speculativ testifies. | ||
| + | |||
| + | * '''Team Activity Tests:''' | ||
| + | ** Activity assay of all 4 laccases and TVEL0 were done at 10 °C. | ||
| + | ** TVEL5 was incubated in copper and after adjusting buffer composition of the supernatant the activity test was done. The result is shown in the following figure: | ||
| + | [[File:Bielefeld2012_TVEL5_Eppis.jpg|thumb|center|350px|Activity test of cultivation (culture 1, 2, 3) supernatant of [http://partsregistry.org/Part:BBa_K863207 BBa_K863207]. The control is the supernatant of ''P. pastoris'' GS115.]] | ||
| + | <br style="clear: both" /> | ||
===Friday October 26th=== | ===Friday October 26th=== | ||
| + | * '''Wiki day''' | ||
| + | * '''Team Cultivation and Purification''' | ||
| + | Final offline analysis of the ''E.coli'' Rosetta Gami 2 cultivations with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09). | ||
| + | |||
===Saturday October 27th=== | ===Saturday October 27th=== | ||
===Sunday October 28th=== | ===Sunday October 28th=== | ||
Latest revision as of 02:40, 27 October 2012
Week 24 (10/08 - 10/14/12)
Contents |
Monday October 08th
All: Day off in Amsterdam!!!
Tuesday October 09th
Wednesday October 10th
- Team Shuttle Vector:
- Ligation of pSB1C3::BBa_K863202 construct in KRX cells was done.
- Team Cellulose Binding Domain:
- Gradient-PCRs of CBDcex_Freiburg; CBDclos_Freiburg; GFP_Freiburg and ecol_Freiburg (47 °C to 67°C)
- Product:
- CBDcex_Freiburg at all temperatures
- CBDclos_Freiburg at all temperatures
- GFP_Freiburg at all temperatures
- ecol_Freiburg at 53 °C to 61°C (best at 57 to 59 °C)
- pooled fractions for gel clean up
- Product:
- Gradient-PCRs of CBDcex_Freiburg; CBDclos_Freiburg; GFP_Freiburg and ecol_Freiburg (47 °C to 67°C)
- Team Site Directed Mutagenesis:
- Ordered Primers for the SDM of the illegal XbaI restriction site in the shuttle vector
- Team Cultivation and Purification:
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]for 19L fermentation (500mL preculture)
Thursday October 11th
- Team Cellulose Binding Domain:
- Clean-Up of the Gel-pieces (from PCR-products)
- Restriction of PCR-products (3h):
- GFP_Freiburg:
- XbaI+PstI Tango-Buffer for Ligation
- XbaI+AgeI Tango 4xAgeI for Assembly
- CBDclos/cex:
- NgoMIV+PstI P.4+BSA 2xPstI for Assembly
- GFP_Freiburg:
- Assembly and Ligation (1h with T4):
- GFP_Freiburg + CBDcex_Freiburg + J61101
- GFP_Freiburg + CBDclos_Freiburg + J61101
- GFP_Freiburg + J61101
- Transformation in E. coli KRX (3 µL, chemical):
- GFP_Freiburg + CBDcex_Freiburg + J61101
- GFP_Freiburg + CBDclos_Freiburg + J61101
- GFP_Freiburg + J61101
- All Transformations were plated on AMP-select-agar
- Team Cultivation and Purification
- 12 L Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]
- Settings:
- 12 L Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]
Bioengineering NFL 19L fermenter, autoinduction HSG medium, 60 µg/mL chloramphenicol, 19 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 12 NL/m, 16 hours. HSG medium was choosen to get a high biomass concentration with hope for a higher amount of laccases.
- Made precultures of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) behind a constitutive promoter. (500mL preculture)
Friday October 12th
- Team Cellulose Binding Domain:
- A lot of colonies on all three dishes (J61101+GFP_Freibug;J61101+GFP_Freibug+CBDcex;J61101+GFP_Freibug+CBDclos)
- no obvious green fluorescence, not even at J61101+GFP
- Maybe the combination of J23100 and J61101 isn't strong enough to express a visible amount of GFP
- Colony-PCR of 16 colonies of every dish
- J61101 + GFP_Freiburg:
- 16/16 positive
- plated two on select-agar
- J61101 + GFP_Freiburg + CBDcex:
- 8/16 positve
- plated three on select-agar
- J61101 + GFP_Freiburg + CBDclos:
- 7/16 positive
- plated three on select-agar
- J61101 + GFP_Freiburg:
- A lot of colonies on all three dishes (J61101+GFP_Freibug;J61101+GFP_Freibug+CBDcex;J61101+GFP_Freibug+CBDclos)
- Team Shuttle Vector:
- The induction with 0.5% (v/v) methanol of P. pastoris GS115 cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] was started.
- Team Substrate Analysis:
- Since we have seen some new peaks after Estradiol treated with Laccases in the LC-MS, we wanted to have more degradation products to do an MS so we used a higher concentradion of Estradiol and Laccase and let them incubate for 66 hours.
- Team Cultivation and Purification
- 12L Cutlivations of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] were harvested and stored at 4°C until purification.
- 12 L Cultivation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09)
- Settings:
- 12 L Cultivation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09)
Bioengineering NFL 19L fermenter, HSG medium, 60 µg/mL chloramphenicol, 300 µg/mL ampicillin, 12 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 12 NL/m, 22-24 hours. HSG medium was chosen to get a high biomass concentration with hope for a higher amount of laccases.
- Team Substrate Analysis:
- Dissolved estradiol, estrone and ethinyl estradiol in 50 % (v/v) acetonitrile with a concentration of 200 µg mL-1
- Creating and measuring of negative controls for estradiol, estrone and ethinyl estradiol. Experimental setup: 2 µg substrate, 110 µL BR-buffer in 200 µL reaction volume. Measured at t0 and after 3 hours.
Saturday October 13th
- Team Shuttle Vector:
- The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were also inducted with 0.5% (v/v) methanol (100%).
- Team Cultivation and Purification
- 12L Cutlivations of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) were harvested and stored at 4°C until purification.

- Team Substrate Analysis:
- Creating and measuring of negative controls for estradiol, estrone and ethinyl estradiol. Experimental setup: 2 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer in 200 µL reaction volume. Measured at t0 and after 10 minutes.
Sunday October 14th
- Team Shuttle Vector:
- The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were also inducted with 0.5% (v/v) methanol (100%).
- Team Fungal and Plant Laccases:
- Team Cellulose Binding Domain:
- Tried to isolate all plates, but only got low concentrations (5 ng/µL - 20 ng/µL)
- plated eight different colonies for isolation
- Team Substrate Analysis:
- We took sample from the reaction of the 12th October.
Week 25 (10/15 - 10/21/12)
Contents |
Monday October 15th
- Team Shuttle Vector:
- There are no colonies of pBS1C3::BBa_K863202 KRX transformantion.
- The P. pastoris GS115 cells integrated with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] were harvested and the supernatant was examined on GFP by fluorescence. But there was no GFP.
- Team Fungal and Plant Laccases:
- Yeah, the yeast cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863207 BBa_K863207] are growing.
- Team Cellulose Binding Domain:
- Tried to isolate the eight plated, but did get low concentrations again (except one J61101+GFP_Freiburg+CBDcex_Freiburg with conc >100 ng/µL)
- cleaned up some more PCR-products (GFP_Freiburg; CBDcex_Freiburg; CBDclos_Freiburg)
- Team Substrate Analysis: We stopped the reaction from the 12th October with Methanol and gave it to Dr. Marcus Persicke to measure the degradation on LC-MS
- Team Cultivation and Purification
- Harvested Cells from 12L Cutlivations of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] as well as 12 L Cultivation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) were disrupted via high-pressure homogenizer and filtrated by [http://www.millipore.com/catalogue/module/C7493 Millipore Pellicon XL 50] with first 300 kDa to seperate all celldebris and 10 kDa to concentrate the laccases. The concentrated solutions were purified by the Ni-NTA-column.
- Team Substrate Analysis:
- Creating and measuring of estradiol and ethinyl estradiol degradation samples for the laccase TVEL0. Experimental setup: 2 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 10 µg laccase in 200 µL reaction volume. Measured at t0 and after 10 minutes.
- Team Immobilization
- Started immobilization of TVEL0 to analyze the binding of laccase during the first 10 hours representative for following immobilizations.
Tuesday October 16th
- Team Activity Tests:
- Today we received the samples from Team Cultivation & Purification. We are really motivated to measure the activity of our new produced enzymes. But before that we have to prepare the samples. This implied the determination of the protein concentration in each fraction, which we did today. The results are listed in the following table:
- Team Cellulose Binding Domain:
- Transformed B0034 into KRX
- Transformed J23100 into KRX
- Plated both on AMP-select-agar
- Plated one more Colony of J61101 + GFP_Freiburg and J61101 + GFP_Freiburg + CBDclos
- Team Substrate Analysis: The Massspectrometry data showed two potential products after the Laccase treatment of Estradiol and Ethinyl estradiol but we unfortunatly could not identify them so we decided to do MS-MS on thoose. The products showed that after Laccase treatment both Estradiol and Ethinyl estradiol losses two H atoms which was new for us.
- Team Immobilization:
Taking samples for immobilization time analyzation.
Wednesday October 17th
- Team Activity Tests:
- Before starting our activity assay we re-buffered the fractions we got from Team Cultivation and Purification. It took some time, that's why we had no time for activity analysis yet. But we prepared everything for the next day, including determining the protein concentration of the fractions after they have been re-buffered. This is what we got:
- Team Shuttle Vector:
- Genomic DNA isolation of yeast cells with integrated [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] construct was done with the Kit. For phenotype characterisation an PCR with the primer pair 5AOX-Phenotype-FW and TT-Phenotype-RV was done and the fragment size was determined. The results are: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863205 BBa_K863205] was not integrated.
- Team Fungal and Plant Laccases:
- YPD liquid culture was inoculated with one yeast colony ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863207 BBa_K863207]) and incubated at 30°C and 150 rpm.
- Team Cellulose Binding Domain:
- Picked colonies of B0034 and J23100 and plated them on AMP selection agar for plasmid isolation
- Team Site Directed Mutagenesis:
- SDM-PCR of the Shuttle vector
- Added DpnI over night
- Team Cultivation and Purification:
- Made SDS-Pages of the cultivations from 10/11 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]) and 10/12 ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012], <partinfo>BBa_K863022</partinfo>).

- Team Immobilization:
Measurement of Protein concentration with Roti-Nanoquant and analyses of binding capacity.
Thursday October 18th
- Team Activity Tests:
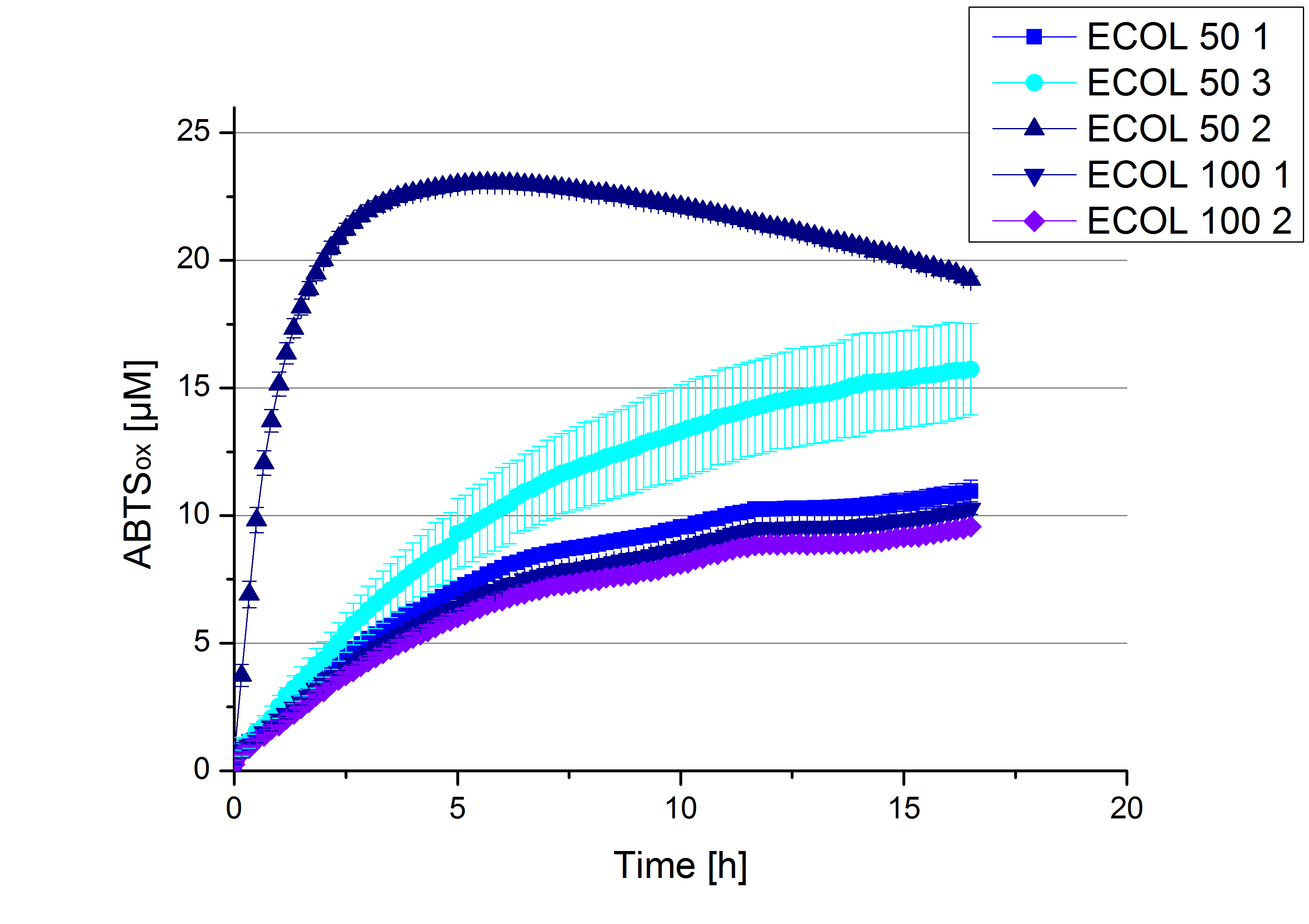
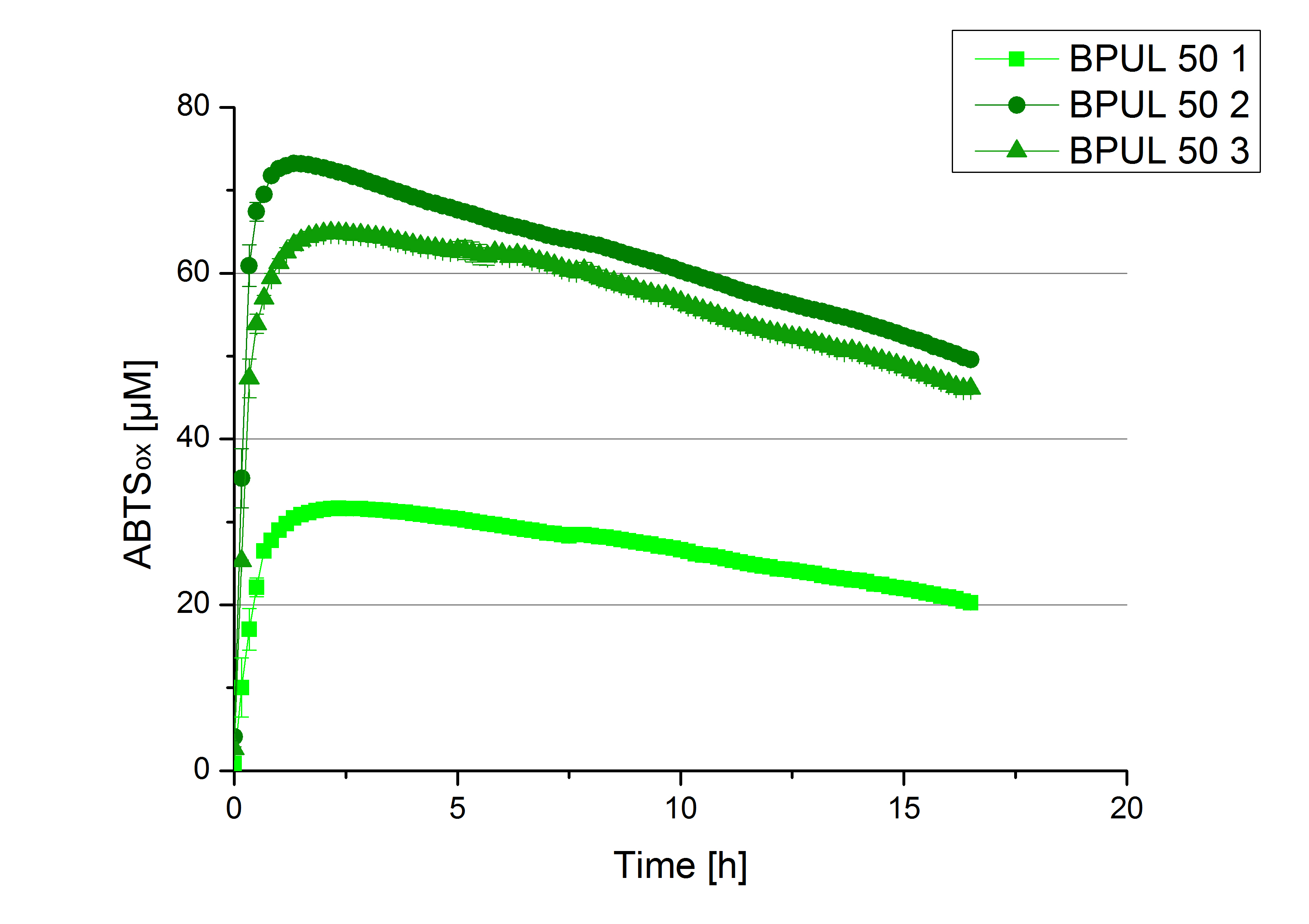
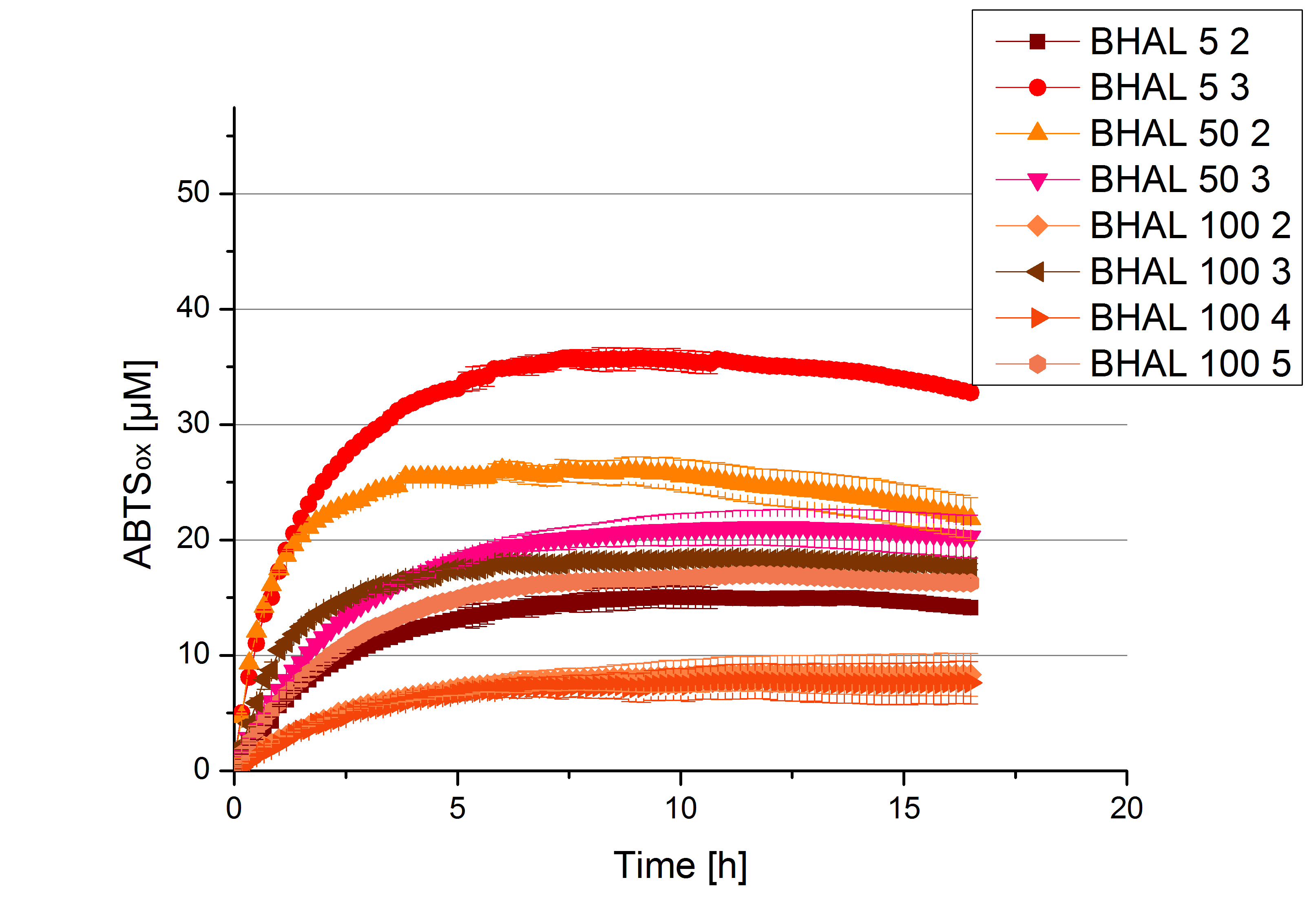
- Finally the day had come to measure the activity of our own produced laccases. This activity assay should give information about the enzyme content in every fraction. To make the measurements comparable we applied the same protein amount in every sample. Again, we used our standard activity assay protocol, but this time we used the Britton-Robinson Buffer at pH 5. Also we applied 0.1 mM ABTS and measured over night.
- Team Cellulose Binding Domain:
- Isolated J23100 plasmid
- Digested J23100 with EcoRI and PstI
- Digested pSB1C3-Backbone with EcoRI and PstI
- Clean up of pSB1C3-BB and J23100 + RFP - Insert via Gel
- Ligation of J23100 and pSB1C3-BB
- Isolated B0034 plasmid
- Digested B0034 with SpeI and PstI
- Digested GFP_Freiburg-PCR-product with XbaI and PstI
- Digested an other fraction of the GFP_Freiburg-PCR-product with XbaI and AgeI
- Assembled (Ligation):
- B0034 with GFP_Freiburg
- B0034 with GFP_Freiburg and CBDcex_Freiburg
- B0034 with GFP_Freiburg and CBDclos_Freiburg
- Transformed these three into KRX
- Team Site Directed Mutagenesis:
- Bands of 19C-PRC-product in Gel at 7 kbp (which is correct)
- Clean-Up of 19C-PRC-product
- Transformation of the 19C-PRC-product in XL1 Blue
- Team Immobilization:
- Discussed new method of activity measurements for immobilized laccases on beads with less lacasse solution needed. Started first general tests with photometer.
Friday October 19th
- Team Activity Tests:
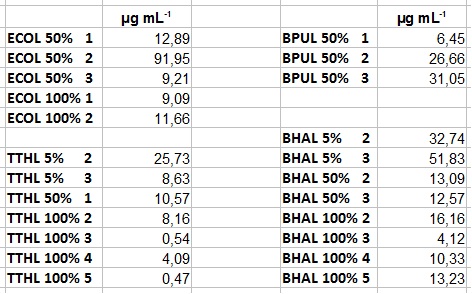
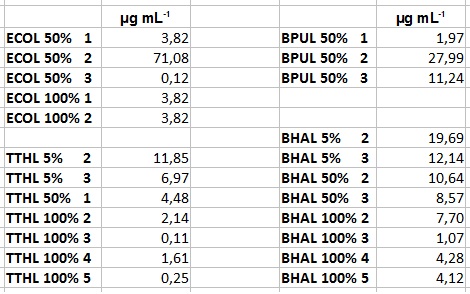
- After getting our interesting activity measurement results, we had to figure out which fraction is going to be used in the following experiments and how much laccase it contains besides other proteins. We agreed, that the most active fraction contains 90 % laccase, which is commonly used in the literature. Since we applied the same protein amount of each fraction for the activity measurements, we made sure that the results really correspond to the amount of laccase. These are the most active fractions we have chosen with their contained laccase amount:
- ECOL: Fraction 50% 2 with a laccase concentration of 63,9 µg mL-1.
- BPUL: Fraction 50% 2 with a laccase concentration of 25,1 µg mL-1.
- BHAL: Fraction 5% 3 with a laccase concentration of 10,9 µg mL-1.
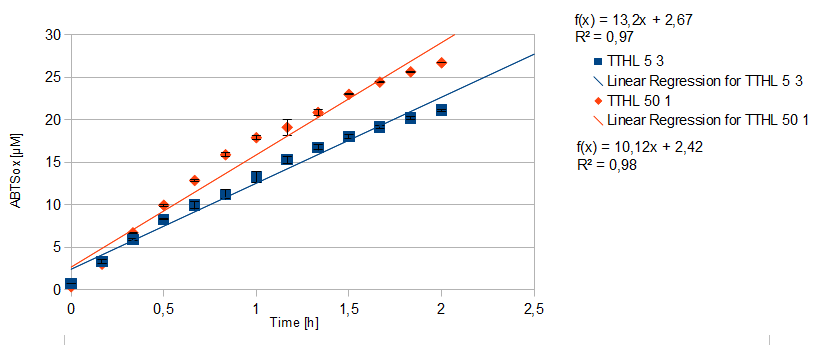
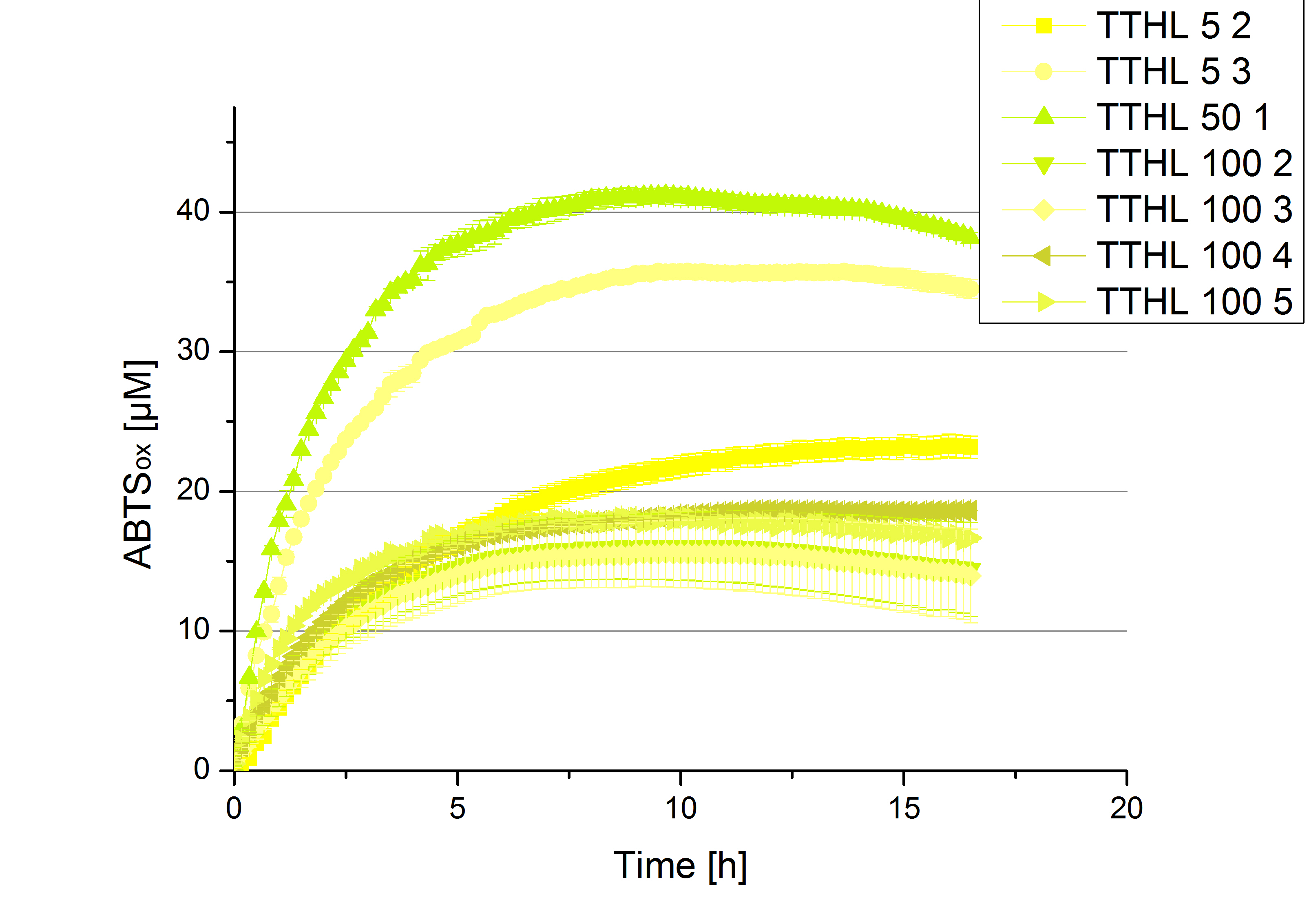
- TTHL: The best fraction of our TTHL laccase was fraction 50% 1 with a laccase concentration of 4,03 µg mL-1. In regard to Team Immobilization and Team Substrate Analysis we had to reach a higher amount of TTHL laccase. So we thought of combining two fractions. We chose the second best fraction of TTHL, which is 5% 3. To calculate the amount of contained laccase we had to compare the activity and reconsidered the slope. Since we stated the fraction with its highest activity as 90 % contained laccase, we calculated the amount of laccase regarding to that one. The following figure shows the slopes we got out of the activity from fraction 50% 1 and fraction 5% 3 (which is the second best). Accordingly to this, fraction 5% 3 contains 68,9% of laccase, which is 4,8 µg mL-1. In total, the composed fraction contains 4,4 µg mL-1.
- After getting our interesting activity measurement results, we had to figure out which fraction is going to be used in the following experiments and how much laccase it contains besides other proteins. We agreed, that the most active fraction contains 90 % laccase, which is commonly used in the literature. Since we applied the same protein amount of each fraction for the activity measurements, we made sure that the results really correspond to the amount of laccase. These are the most active fractions we have chosen with their contained laccase amount:
- Team Cellulose Binding Domain:
- Colony-PCR of B0034 + GFP_Freiburg; B0034 + GFP_Freiburg + CBDcex; B0034 + GFP_Freiburg + CBDcex_Freiburg; B0034 + GFP_Freiburg + CBDclos_Freiburg.
- All negative
- Restriction of Ecol_Freiburg with XbaI, AgeI and DpnI
- Colony-PCR of B0034 + GFP_Freiburg; B0034 + GFP_Freiburg + CBDcex; B0034 + GFP_Freiburg + CBDcex_Freiburg; B0034 + GFP_Freiburg + CBDclos_Freiburg.
- Team Site Directed Mutagenesis:
- Team Substrate Analysis:
- Creating and measuring of estradiol and ethinyl estradiol degradation samples for the laccases TVEL0 and BPUL. The experimental setup changed. The laccase amount was adjusted to the highest possible amount of the lowest concentrated laccase. TTHL has a concentration of 4,03 µg mL-1 and sets the maximal laccase amount to 0.6 µg per 200 µL reaction volume. To compensate these worse conditions, the substrate amount was lowered to 1 µg and the temperature was changed to 30°C.
- Test of estradiol and ethinyl estradiol degradation done by TVEL0 and BPUL without ABTS. Experimental setup: 1 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 0.6 µg laccase in 200 µL reaction volume. Measured at t0 and after 3 hours.
Saturday October 20th
- Team Activity Tests:
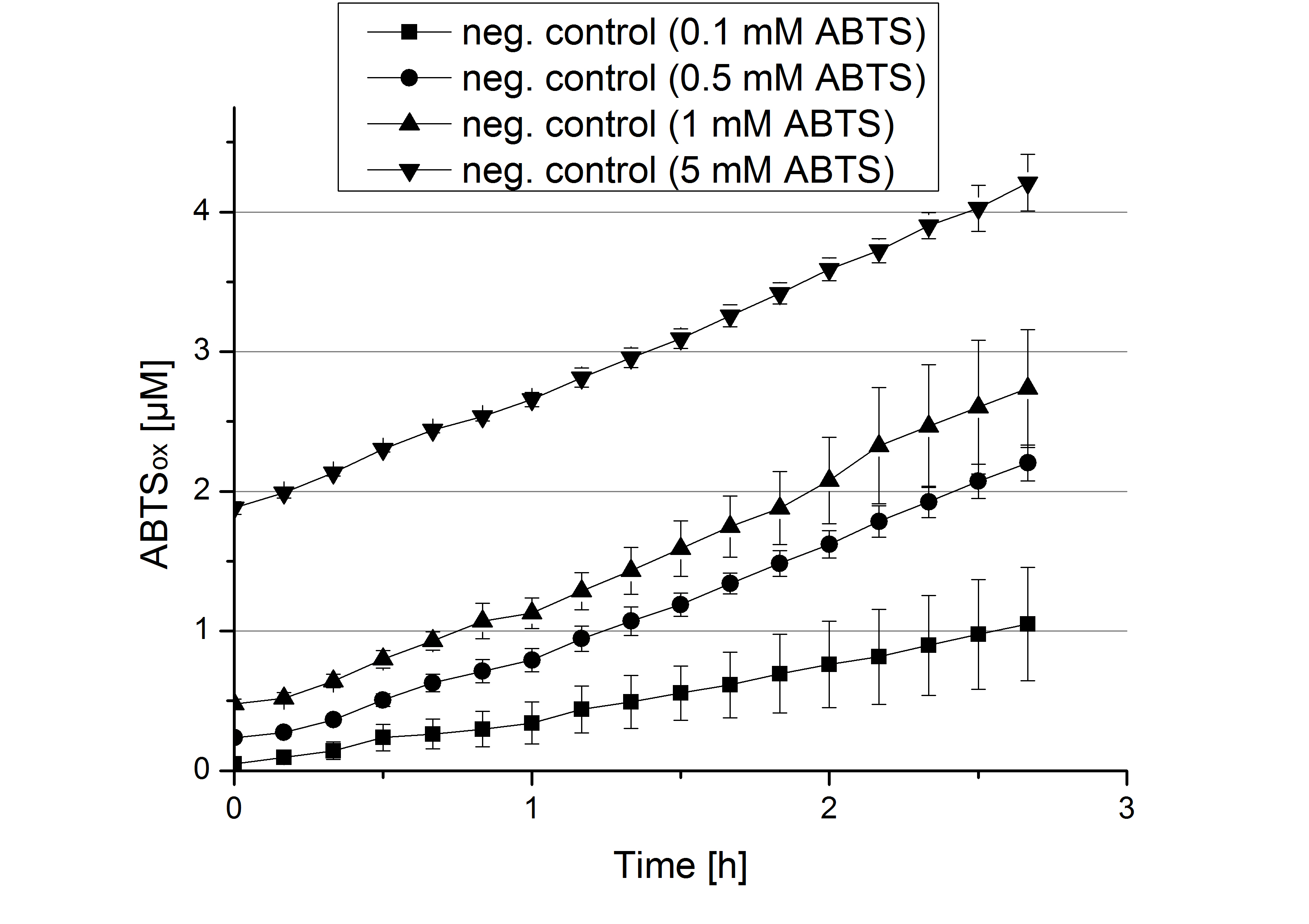
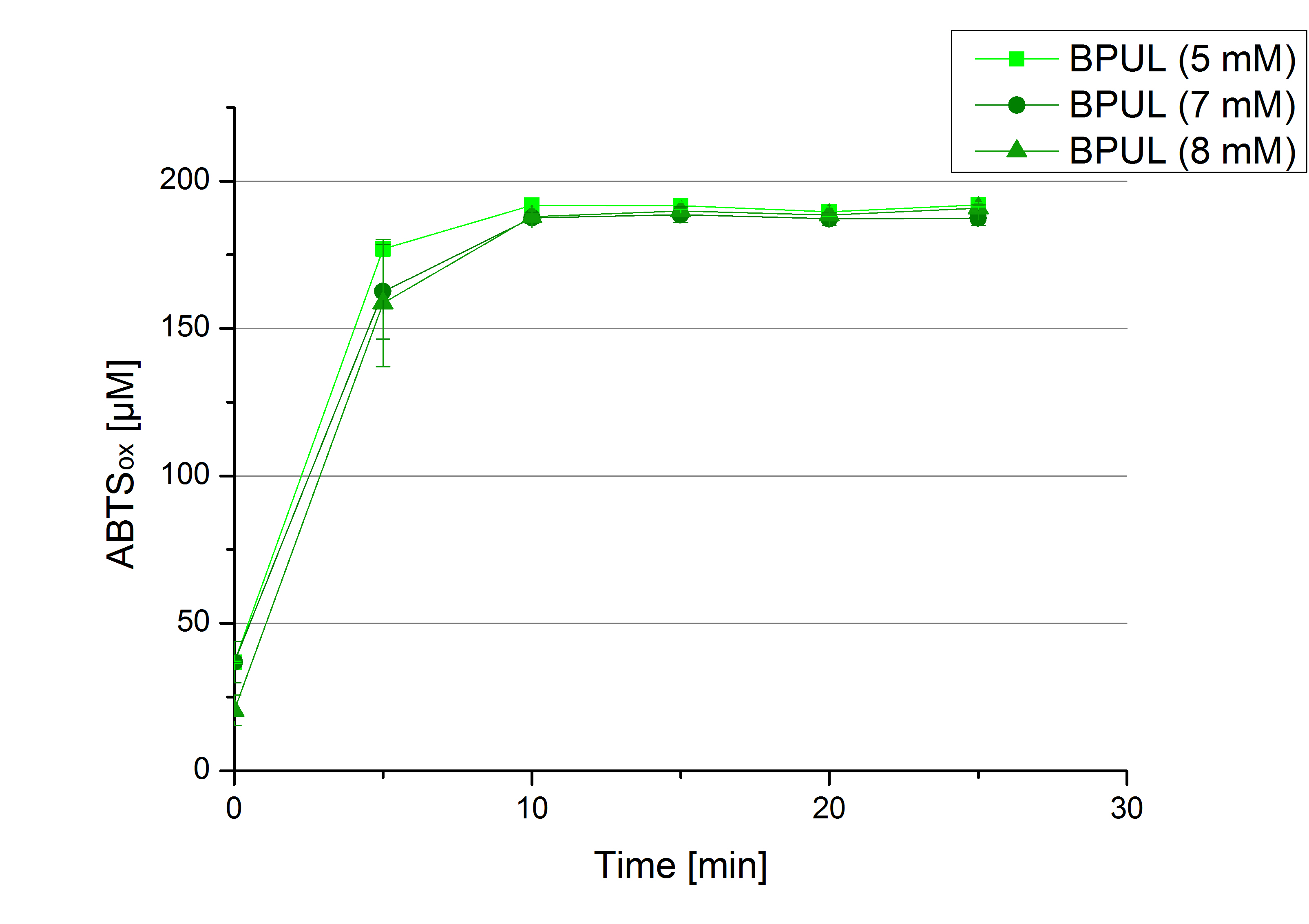
- During the Jamboree in Amsterdam we had the oppurtunity to discuss our activity results with a lot of people. One remark was about the calculation of the enzyme specific units. As you all remember, we measured the activty with 0.1 mM ABTS, but to get comparable results with the literature, we had to measure each protein with other concentrations of ABTS. The concentration of ABTS should be saturated for each laccase. So we started with ECOL and BPUL and applied different ABTS concentrations starting from 0.1 mM as usual: 0.1 mM, 0.5 mM, 1 mM and 5 mM. We applied 616 ng Laccase in each sample. Our results showed, that especially ECOL does not reach its substrate saturation with 5 mM ABTS. Our plans are now to check these concentrations of ABTS for BPUL and TTHL and also higher concentrations of ABTS for all laccases. For that we should reduce the enzyme amount to be able to detect the change in OD 420 properly.
- Team Substrate Analysis:
- TVEL0 and BPUL were able to degrade 100 % of estradiol and 100 % of ethinyl estradiol. To get comparable results, the reaction temperature was lowered to 20 °C again, to have a closer look, where differences are apparent. Again TVEL0 and BPUL degraded 100 % of the substrates. But before the laccase stocks deplete, first ECOL, BHAL and TTHL should be tested.
- Did substrate degradation analysis with ECOL without ABTS. Experimental setup: 1 µg substrate, 0.1 mM ABTS, 110 µL BR-buffer and 0.6 µg laccase in 200 µL reaction volume. Measured at t-0 and after 3 hours.
Sunday October 21st
- Team Fungal and Plant Laccases:
Genomic isolation was done with the [http://www.promega.com/resources/protocols/technical-manuals/0/wizard-genomic-dna-purification-kit-protocol/ Promega Wizard genomic DNA purification system kit] of our approximately 50 yeast clones with integrated (a) GFP and (b) tvel5.
- Team Activity Tests:
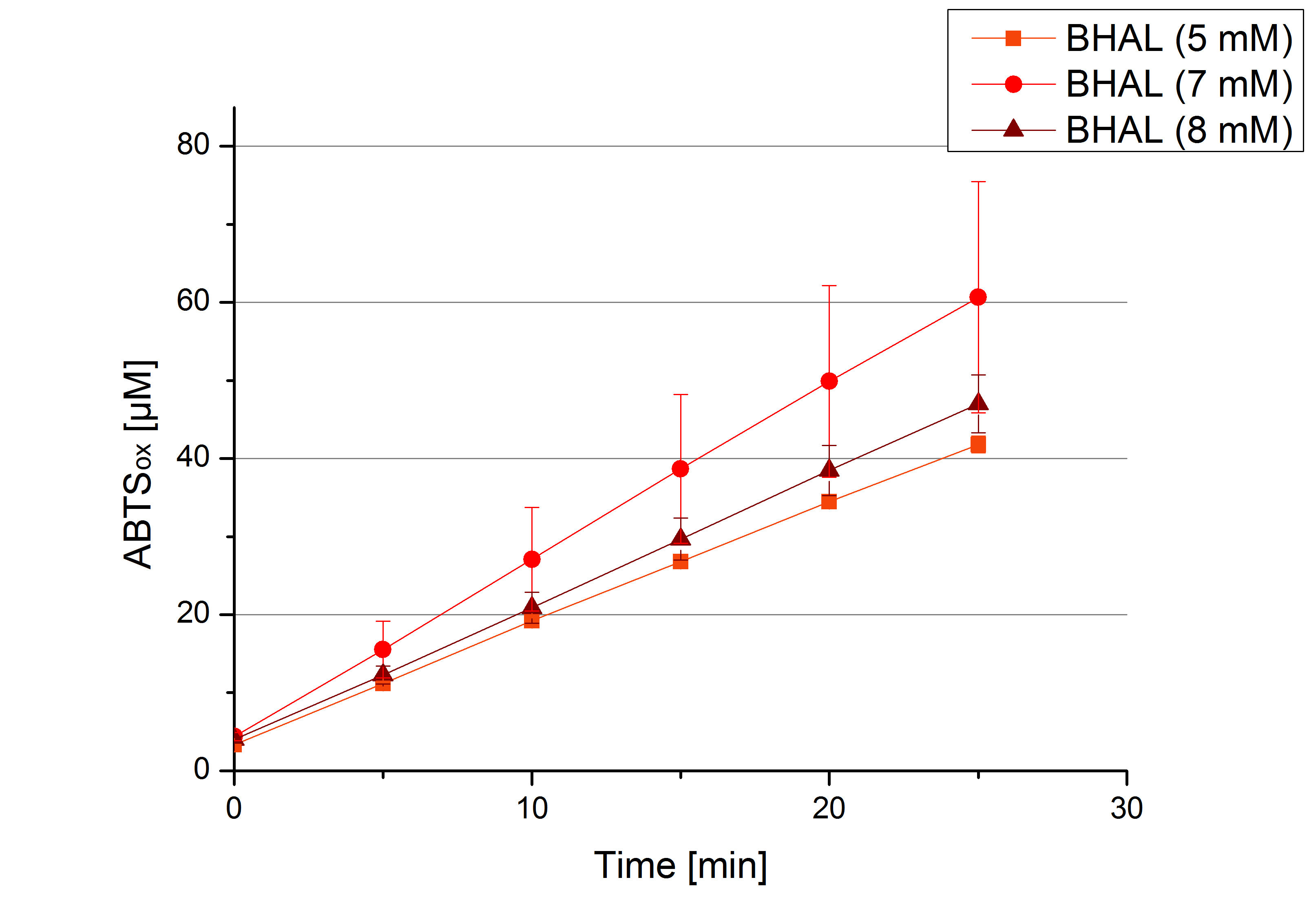
- Today was BHAL's and TTHL's turn. The experiment setup was the same as described the day before. But again we could not make sure, that 5 mM ABTS is the substrate saturation for our tested laccases. We are now sure to go higher in ABTS concentrations.
- Team Cellulose Binding Domain:
- Colonies PCRs of B0034 + GFP_Freiburg; B0034 + GFP_Freiburg + CBDcex_Freiburg; B0034 + GFP_Freiburg + CBDclos_Freiburg
- All negativ
- Transformed once again (with an mix of old an new GFP_Freiburg)
- Isolated plated colonies of transformed colonies (two GFP; two GFP+Cex; two GFP+Clos
- Restriction analysis showed only bands at 2K bp (pSB1C3+BBa_B0034 vector without insert)
- Colonies PCRs of B0034 + GFP_Freiburg; B0034 + GFP_Freiburg + CBDcex_Freiburg; B0034 + GFP_Freiburg + CBDclos_Freiburg
- Team Site Directed Mutagenesis:
- Isolated the plasmids of the six SDM-dishes
- Over-night digestion with NotI
- Team Substrate Analysis:
- ECOL, BHAL and TTHL were tested with the same conditions and all of them degraded the substrates much slower than TVEL0 and BPUL.
- Team Immobilization
- Started immobilization of new laccases ECOL, BPUL, BHAL, TTHL and TVEL0.
Week 26 (10/22 - 10/28/12)
Contents |
Monday October 22nd
- Team Fungal and Plant Laccases:
The identification of positive yeast clones and determination of the phenotype (M+ or MS) was done by PCR with the primers 5AOX-Phenotype-FW and TT-Phenotype-RV. The result of the PCR was: no GFP integrated clones and three tvel5 integrated (1.9 kb and 2.2 kb) clones. The cassette, including tvel5 and his4, was recombinated by a single cross over, because the 2.2 kb PCR product of the aox1 gene has been formed. Minimal methanol medium was inoculated with these three positive clones with the M+ phenotype.
- Team Activity Tests:
- Today was all about finding a comfy environment for our laccases. We thought of determining the right pH before going on with substrate saturation experiments since we could not be sure, that the activity was influenced by the pH. We chose a range from pH 4 to pH 9, applied 616 ng laccase and 5 mM ABTS. Our results revealed the following results:
- optimal pH of ECOL: pH 5
- optimal pH of BPUL: pH 4 and pH 5
- optimal pH of BHAL: pH 5
- optimal pH of TTHL: pH 5
-
- Team Cellulose Binding Domain:
- Sequencing of <partinfo>BBa_K863120</partinfo> in <partinfo>BBa_J61101</partinfo> showed a deletion within the ORF of the GFP; this seemed to be the reason that the colonies are not fluorescent.
- Gradient-PCR on <partinfo>BBa_I13522</partinfo> with GFP_Freiburg Pre- and Suffix Primers
- Worked fine. Clean up and digestion with XbaI and PstI
- Gradient-PCR on <partinfo>BBa_K863103</partinfo> with CBDcex_Freiburg and GFP_Freiburg_compl Primers
- One of 12 temperatures worked. Clean up and digestion with XbaI and PstI
- Gradient-PCR on <partinfo>BBa_K863113</partinfo> with CBDclos_Freiburg and GFP_Freiburg_compl Primers
- did not get product at all
- Ligation of the PCR products and GFP_Freiburg and CBDcex_Freiburg-GFP_Freiburg, respectively
- Transformation of both ligations
- Cells plated on selection agar
- Team Site Directed Mutagenesis:
- Stopped Digestion of the shuttle vector, but gel went wrong and the products were lost
- Team Substrate Analysis:
- BHAL and TTHL were tested with the same conditions and all of them degraded the substrates much slower than TVEL0 and BPUL.
- Team Immobilization:
- Measured of protein concentration in supernatants of immobilized ECOL, BPUL, BHAL, TTHL and TVEL0 with Roti-Nanoquant. Analysed binding capacity of the different laccases.
Tuesday October 23rd
- Team Fungal and Plant Laccases: The induction of the yeast cells with 1% (v/v) 100% methanol was done at morning and in the evening.
- Team Activity Tests:
- In order to find the substrate saturation we continued to measure our laccase activities with higher concentrations of ABTS. To have a slower reaction and also try not to waste the enzymes we halfed the amount of used laccases from 616 ng to 308 ng of each laccase. The following substrate saturations were determined:
- ECOL: 9 mM ABTS
- BPUL: 5 mM ABTS
- BHAL: 7 mM ABTS
- TTHL: 7 mM ABTS
- In order to find the substrate saturation we continued to measure our laccase activities with higher concentrations of ABTS. To have a slower reaction and also try not to waste the enzymes we halfed the amount of used laccases from 616 ng to 308 ng of each laccase. The following substrate saturations were determined:
- Team Cellulose Binding Domain:
- Colony PCR of B0034 + GFP_Freiburg
- 14/24 positive
- Plated three on selection agar for plasmid isolation
- Colony PCR of B0034 + CBDcex-GFP_Freiburg
- 16/24 positive
- Plated three on selection agar for plasmid isolation
- Colony PCR of B0034 + GFP_Freiburg
- Team Immobilization
- Started measurement of enzymatic activity of the different immobilized laccases with the Photometer.
Wednesday October 24th
- Team Activity Tests:
- pH measurements were repeated under substrate saturation with a pH range from pH 4 to pH 9.
- Team Fungal and Plant Laccases: In the morning and evening the induction of yeast cells with 1% (v/v) methanol was done.
- Team Cellulose Binding Domain:
- Isolated Plasmids containing B0034 + GFP_Freiburg &
- Digestion of pooled plasmids with XbaI and PstI
- Isolated Plasmids containing B0034 + CBDcex-GFP_Freiburg
- Digestion of pooled plasmids with XbaI and PstI
- Digestion of J23100 with SpeI and PstI
- Clean up of inserts (B0034 + GFP_Freiburg & B0034 + CBDcex-GFP_Freiburg) and Backbone (J23100) via gel
- Ligation of Backbone and the inserts
- Transformation and plated on selection agar
- Isolated Plasmids containing B0034 + GFP_Freiburg &
- Team Site Directed Mutagenesis:
- Digestion showed that all isolated colonies still have the illegal XbaI restriction side
- Team Substrate Analysis:
- Repeating of the degradation experiments without ABTS for TVEL0, BPUL, ECOL, BHAL and TTHL with a degradation time of five hours to increase the differences between the laccases.
Thursday October 25th
- Team Fungal and Plant Laccases:
- Induction of the yeast cells with 1% (v/v) methanol was done. After 4 h the cells were harvested by centrifugation at 3000xg 4°C for 5 min.
- Team Cellulose Binding Domain:
- All colonies did not show any obvious green glow
- Team Substrate Anaylsis:
- The MS-MS data arrived from Marcus Persicke. To ensure our measurements and discuss what happens after the Laccase treatment and on the ESI we decided to ask an organic chemistrer so we went to Prof. Dr. Dietmar Kuck whos expert on this field. Since he had less time to discuss we have only speculativ testifies.
- Team Cultivation and Purification:
Last offline analysis of the the two cultivations of E.coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] with Carobon Source Detection with HPLC.
- Team Substrate Analysis:
- Repeating of the degradation experiments with ABTS for ECOL, BHAL and TTHL to replace previous experimental errors.
Thursday October 25th
- Team Fungal and Plant Laccases:
- Induction of the yeast cells with 1% (v/v) methanol was done. After 4 h the cells were harvested by centrifugation at 3000xg 4°C for 5 min.
- Team Cellulose Binding Domain:
- All colonies did not show any obvious green glow
- Team Substrate Anaylsis:
- The MS-MS data arrived from Marcus Persicke. To ensure our measurements and discuss what happens after the Laccase treatment and on the ESI we decided to ask an organic chemistrer so we went to Prof. Dr. Dietmar Kuck whos expert on this field. Since he had less time to discuss we have only speculativ testifies.
- Team Activity Tests:
- Activity assay of all 4 laccases and TVEL0 were done at 10 °C.
- TVEL5 was incubated in copper and after adjusting buffer composition of the supernatant the activity test was done. The result is shown in the following figure:
Friday October 26th
- Wiki day
- Team Cultivation and Purification
Final offline analysis of the E.coli Rosetta Gami 2 cultivations with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09).
Saturday October 27th
Sunday October 28th
| 55px | | | | | | | | | | |
 "
"