Team:Paris Bettencourt/Overview
From 2012.igem.org
(→Objectives) |
Aleksandra (Talk | contribs) (→Evaluation) |
||
| (109 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/header}} | {{:Team:Paris_Bettencourt/header}} | ||
| - | |||
<div id="grouptitle">Project Overview</div> | <div id="grouptitle">Project Overview</div> | ||
| + | <br> | ||
| + | <html> | ||
| + | <style type="text/css"> | ||
| + | .gallerycontainer{ | ||
| + | position: relative; | ||
| + | /*Add a height attribute and set to largest image's height to prevent overlaying*/ | ||
| + | } | ||
| + | |||
| + | .thumbnail img { | ||
| + | border: 1px solid white; | ||
| + | margin: 0 5px 5px 0; | ||
| + | position: relative; | ||
| + | top: -50px; | ||
| + | left: 50px; | ||
| + | } | ||
| + | #txtOV { | ||
| + | left: 598px; | ||
| + | position: relative; | ||
| + | top: -555px; | ||
| + | width: 400px; | ||
| + | } | ||
| + | |||
| + | .thumbnail:hover{ | ||
| + | background-color: transparent; | ||
| + | } | ||
| + | /* | ||
| + | .thumbnail:hover img{ | ||
| + | border: 1px solid blue; | ||
| + | } | ||
| + | */ | ||
| + | .thumbnail span { /*CSS for enlarged image*/ | ||
| + | position: absolute; | ||
| + | top : 80px; | ||
| + | border: none; | ||
| + | visibility: hidden; | ||
| + | color: black; | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | .thumbnail span img{ /*CSS for enlarged image*/ | ||
| + | border-width: 0; | ||
| + | padding: 2px; | ||
| + | } | ||
| + | |||
| + | .thumbnail:hover span{ /*CSS for enlarged image*/ | ||
| + | visibility: visible; | ||
| + | top: 60 px; | ||
| + | left: -70px; /*position where enlarged image should offset horizontally */ | ||
| + | z-index: 50; | ||
| + | } | ||
| + | |||
| + | |||
| + | </style> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2012/1/18/TotalBiosafety.png" width="500px" style="left: 32px;position: absolute;top: 443px;"/> | ||
| + | |||
| + | <div class="gallerycontainer"> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="https://static.igem.org/mediawiki/2012/1/1b/MOUSECURSORPB12.png" width="60px" border="0" /><span></span></a> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="https://static.igem.org/mediawiki/2012/e/e3/PhysicalContainment.png" width="60px" border="0" /><span><img src="https://static.igem.org/mediawiki/2012/f/f6/PhysicalContainmentSystemLarge.png" width="500px" /><br /><div id="txtOV"><b>1) Physical containment: Our cells will be encapsulated with Arabinose and LB in order to avoid release of DNA/cells and to permit growth</b> </div></span></a> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="https://static.igem.org/mediawiki/2012/1/1c/SemanticContainment.png" width="60px" border="0" /><span><img src="https://static.igem.org/mediawiki/2012/0/09/SemanticContainmentSystemLarge.png" width="500px" /><br /><div id="txtOV">1) Physical containment: Our cells will be encapsulated with Arabinose and LB in order to avoid release of DNA/cells, while permitting their growth.<br><br><b>2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations. </b></div></span></a> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="/wiki/images/6/6f/DelaySystem.png" width="60px" border="0" /><span><img src="/wiki/images/b/b9/Delay1PB12.gif" width="500px" /><br /><div id="txtOV"> | ||
| + | 1) Physical containment: Our cells will be encapsulated with Arabinose and LB in order to avoid release of DNA/cells, while permitting their growth.<br><br>2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations. <br><br> | ||
| + | <b>3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.</b> | ||
| + | </div></span></a> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="/wiki/images/5/5c/RestrictionSystem.png" width="60px" border="0" /><span><img src="/wiki/images/9/97/Restriction2PB12.gif" width="500px" /><br /><div id="txtOV"> | ||
| + | 1) Physical containment: Our cells will be encapsulated with Arabinose and LB in order to avoid release of DNA/cells, while permitting their growth.<br><br>2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations. <br><br> | ||
| + | 3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.<br><br> | ||
| + | <b>4) Restriction Enzyme system: The restriction enzyme destroys the plasmid carrying the synthetic circuit and the anti-toxin gene.</b></div></span></a> | ||
| + | |||
| + | <a class="thumbnail" href="#thumb"><img src="https://static.igem.org/mediawiki/2012/d/da/SkullIcon.png" width="60px" border="0" /><span><img src="/wiki/images/8/8f/Toxin3aPB12.gif" width="500px"/><br /><div id="txtOV">1) Physical containment: Our cells will be encapsulated with Arabinose and LB in order to avoid release of DNA/cells, while permitting their growth.<br><br>2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations. <br><br> | ||
| + | 3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.<br><br> | ||
| + | 4) Restriction Enzyme system: The restriction enzyme destroys the plasmid carrying the synthetic circuit and the anti-toxin gene.<br><br><b>5) Suicide system: Once the anti-toxin concentration is below a given threshold, the toxin is no longer inhibited. It kills the cell as well as its neighbors, and eliminates extracellular DNA via its DNase activity.</b></div></span></a> | ||
| + | |||
| + | |||
| + | |||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | <!-- ########## Don't edit above ########## --> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | ==Our project - a hypothetical case study== | ||
| + | |||
| + | Imagine a farmer that wants to measure the nutrient concentration in her field, in order to optimize her fertilizer use. We could provide her with cells carrying a nitrate biosensor (AgrEcoli), encapsulated in alginate beads. She would spread the beads in her field, wait for 12 hours, and then check if they are glowing in response to the nitrates in the soil. | ||
| + | |||
| + | We want this system to work the way the original designers intended. We also want to reduce the chance that the engineered bacteria will survive in the soil, release intact DNA, or transfer genes to a soil microbe. | ||
| + | |||
| + | Our system will be implemented as a pair of plasmids, compatible with the most commonly used BioBrick plasmids, and therefore easily integrated with existing systems. The AgrEColi are now running bWARE, and express a new containment functionality. | ||
| + | |||
| + | Shortly after the beads enter the soil, the delay system triggers and the DNA-degrading colicin proteins become active. The colicins specifically recognize and enter E. coli cells, killing them by completely degrading their genomes. The colicins also degrade DNA loose in the environment, without harming the native bacterial species. | ||
| + | |||
| + | Every field is a different ecosystem with a different composition of native species. The potential consequences of horizontal gene transfer are therefore difficult to predict in general. By destroying the information contained in DNA, our system reduces the chance that introduced DNA will replicate in the new environment. | ||
| + | |||
==Objectives== | ==Objectives== | ||
| Line 8: | Line 108: | ||
*Raise the issue of biosafety, and advocate the discerning use of biosafety circuits in future iGEM projects as a requirement | *Raise the issue of biosafety, and advocate the discerning use of biosafety circuits in future iGEM projects as a requirement | ||
| - | *Evaluate the risk of HGT in different SynBio applications | + | *Evaluate the risk of HGT in different SynBio applications, and perform a fault tree analysis for our project as an example |
*Develop a new, improved containment system to expand the range of environments where GEOs can be used safely. | *Develop a new, improved containment system to expand the range of environments where GEOs can be used safely. | ||
| Line 26: | Line 126: | ||
<tr> | <tr> | ||
<td> | <td> | ||
| - | ==== | + | ====Key Safety Device Design Features==== |
| + | |||
| + | *The physical capsule prevents Genetically Engineered Bacteria (GEB) from escaping into the environment. | ||
| + | *The DNAse toxin kills cells and destroys genetic information. | ||
| + | *A population-level mechanism compensates system failure in single cells. | ||
| + | *Plasmid destruction with restriction enzymes is an irreversible trigger. | ||
| + | *A specific toxin targeting mechanism reduces system impact on native fauna. | ||
| + | *In case HGT occurs, semantic containment prevents the expression of genes from the GEB. | ||
| + | *The modular design supports integration with existing iGEM projects. | ||
| + | *The use of several redundant modules compensates the failure of one of them. | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | ==Evaluation== | ||
| + | <div id="boston"> | ||
| + | It is essential not only to implement a safety system, but also to evaluate its efficiency. | ||
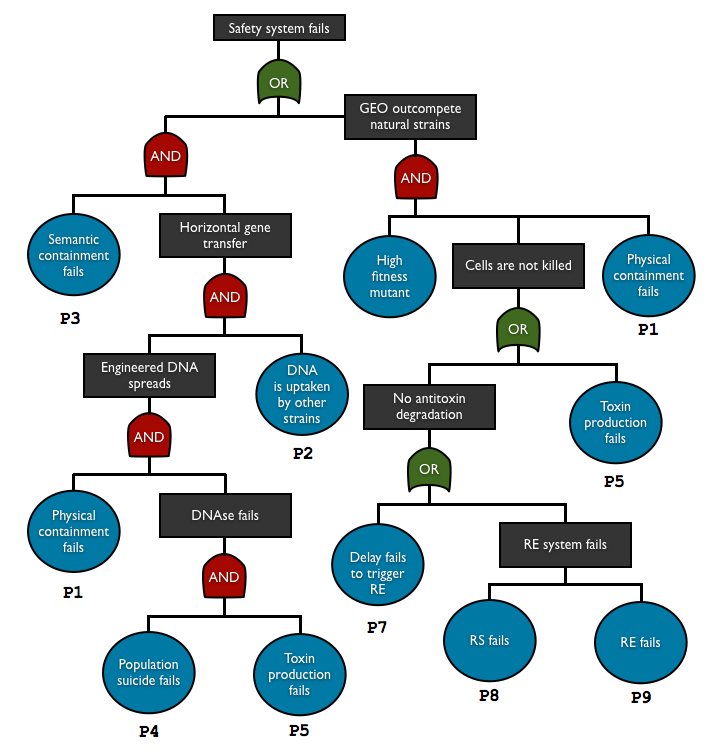
| + | We have addressed this issue by creating a fault tree analysis for our system, but we believe that such approach should be recommended for containment system. You can find more about it on our [https://2012.igem.org/Team:Paris_Bettencourt/Modeling Safety Assessment page]. | ||
| + | <center>[[Image:ParisB_FTA.png]]</center> | ||
| + | </div> | ||
<!-- ########## Don't edit below ########## --> | <!-- ########## Don't edit below ########## --> | ||
{{:Team:Paris_Bettencourt/footer}} | {{:Team:Paris_Bettencourt/footer}} | ||
Latest revision as of 22:52, 26 October 2012






2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations.


2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations.
3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.


2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations.
3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.
4) Restriction Enzyme system: The restriction enzyme destroys the plasmid carrying the synthetic circuit and the anti-toxin gene.

2) Semantic Containment: Our synthetic system will have an amber codon embedded in its genes. The amber suppressor system will ensure their expression in our bacteria, while preventing it in natural populations.
3) Delay system: In the presence of Arabinose, LacI is produced, repressing the expression of a restriction enzyme. Once Arabinose is not present any more, the LacI repressor concentration decreases with dilution and degradation. This leads to the expression of the restriction enzyme.
4) Restriction Enzyme system: The restriction enzyme destroys the plasmid carrying the synthetic circuit and the anti-toxin gene.
5) Suicide system: Once the anti-toxin concentration is below a given threshold, the toxin is no longer inhibited. It kills the cell as well as its neighbors, and eliminates extracellular DNA via its DNase activity.
Contents |
Our project - a hypothetical case study
Imagine a farmer that wants to measure the nutrient concentration in her field, in order to optimize her fertilizer use. We could provide her with cells carrying a nitrate biosensor (AgrEcoli), encapsulated in alginate beads. She would spread the beads in her field, wait for 12 hours, and then check if they are glowing in response to the nitrates in the soil.
We want this system to work the way the original designers intended. We also want to reduce the chance that the engineered bacteria will survive in the soil, release intact DNA, or transfer genes to a soil microbe.
Our system will be implemented as a pair of plasmids, compatible with the most commonly used BioBrick plasmids, and therefore easily integrated with existing systems. The AgrEColi are now running bWARE, and express a new containment functionality.
Shortly after the beads enter the soil, the delay system triggers and the DNA-degrading colicin proteins become active. The colicins specifically recognize and enter E. coli cells, killing them by completely degrading their genomes. The colicins also degrade DNA loose in the environment, without harming the native bacterial species.
Every field is a different ecosystem with a different composition of native species. The potential consequences of horizontal gene transfer are therefore difficult to predict in general. By destroying the information contained in DNA, our system reduces the chance that introduced DNA will replicate in the new environment.
Objectives
Our project aims to:
- Raise the issue of biosafety, and advocate the discerning use of biosafety circuits in future iGEM projects as a requirement
- Evaluate the risk of HGT in different SynBio applications, and perform a fault tree analysis for our project as an example
- Develop a new, improved containment system to expand the range of environments where GEOs can be used safely.
To do so, we:
- Engaged the general public and scientific community through debate
- Raised the question about how we can regulate this practices
- Compiled a parts page of safety circuits in the registry
- Relied on three levels of containment :
- Physical containment with alginate capsules
- Semantic containment using an amber suppressor system
- An improved killswitch featuring delayed population-level suicide through complete genome degradation.
We strived to make our system as robust against mutations as possible.
Key Safety Device Design Features
|
Evaluation
It is essential not only to implement a safety system, but also to evaluate its efficiency.
We have addressed this issue by creating a fault tree analysis for our system, but we believe that such approach should be recommended for containment system. You can find more about it on our Safety Assessment page.
 "
"


 Overview
Overview Delay system
Delay system Restriction enzyme system
Restriction enzyme system MAGE
MAGE Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment