Team:HokkaidoU Japan/Notebook/Week 3
From 2012.igem.org
| Line 181: | Line 181: | ||
===Liquid Culture=== | ===Liquid Culture=== | ||
Liquid culture for single colony isolated pT7-RBS on pSB1C3 and Ag43-dT on pSB1T3. | Liquid culture for single colony isolated pT7-RBS on pSB1C3 and Ag43-dT on pSB1T3. | ||
| - | #Picked up one colony from single colony isolated plates by platinum loop. | + | |
| + | '''Ag43-dT on pSB1T3 colonies were closely existed so we would picked up two or more colonies.''' | ||
| + | #Picked up one (or tow?) colony from single colony isolated plates by platinum loop. | ||
#Dipped into 2ml of LBC and LBT. | #Dipped into 2ml of LBC and LBT. | ||
#Cultivated. | #Cultivated. | ||
| Line 203: | Line 205: | ||
If digestion were succeeded, there are two bands would be seen: about 3120bp and 2070bp. | If digestion were succeeded, there are two bands would be seen: about 3120bp and 2070bp. | ||
| - | And if mini-prep were succeeded, there would | + | And if ligation-transformation-mini-prep were succeeded, there would be existed two bands in each lanes: about 5000bp and 2000bp. |
[[image:HokkaidoU2012 120717 Ag43-Ag43d+(E&S)-(Ag43-dT on pSB1T3)-(pT7-RBS on pSB1C3).jpg|thumb|Digestion and mini-prep result image]] | [[image:HokkaidoU2012 120717 Ag43-Ag43d+(E&S)-(Ag43-dT on pSB1T3)-(pT7-RBS on pSB1C3).jpg|thumb|Digestion and mini-prep result image]] | ||
| Line 210: | Line 212: | ||
| - | About mini-prep products, | + | About mini-prep products, in Ag43-dT on pSB1T3, there were two plasmid DNA bands about 8000bp and 3000bp in one lane. We thought '''which means we failed single colony isolation then resuspended two another E.coli colonies''' another ligated DNA were transformed. |
| + | In pT7-RBS on pSB1C3, we needed about 2000bp plasmid DNA and there were about 1500bp of DNA. Plasmid DNA migrates more far than linear DNA so we thought we got correct DNA. | ||
| + | |||
</div> | </div> | ||
<!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT UNDER THIS LINE @iTakeshi --> | ||
Revision as of 07:47, 17 July 2012
Contents |
July 16th
Ag43, dT
Digestion
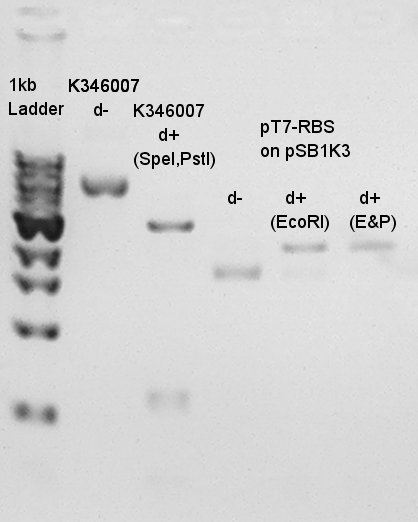
Results of digestion in 15th.
Product:Ag43(K346007)=3120bp and 2070bp, pT7-RBS on pSB1K3=2247bp We confirmed there are some kind of restriction enzyme site in K346007 (digested with SpeI, PstI) and pT7-RBS on pSB1K3 was successfully digested with EcoRI and PstI. Balance between d+(EcoRI) and d+(E&P:cut with EcoRI and PstI) is about 80bp.
Gel Extraction
Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit(NipponGenetics). Got 50ul of DNA solution.
Ethanol Precipitation
Ethanol Precipitation for digestion and gel extraction products.
- Added 5ul of NaoAc, 1.5ul of glycogen and 125ul of 100% ethanol.
- Centrifuged in 15000rpm, 10min at 4C.
- Remove supernatant and added 220ul of 70% ethanol.
- Centrifuged in 15000rpm, 10min at 4C.
- Remove supernatant and air drying in room temperature then added 5ul of DW.
dT(B0015) would be amplified incorrectly and couldn't get enough digestion product. So we tried another DNA amplification method: PCR then digested.
PCR
PCR for dT(B0015)
| DNA solution | 1ul |
| KOD-Plus-NEO(Taq polymerase) | 1ul |
| dNTP | 5ul |
| MgSO4 | 3ul |
| KOD-Plus-NEO Buffer | 5ul |
| Forward Primer(100bp_up forward primer) | 1ul |
| Reverse Primer(200bp_down Reverse primer) | 1ul |
| DW | 33ul |
| Total | 50ul |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58 | 30 |
| 4 | 68 | 30 |
| 5 | 4 | HOLD |
Cycle:2~4 x 45
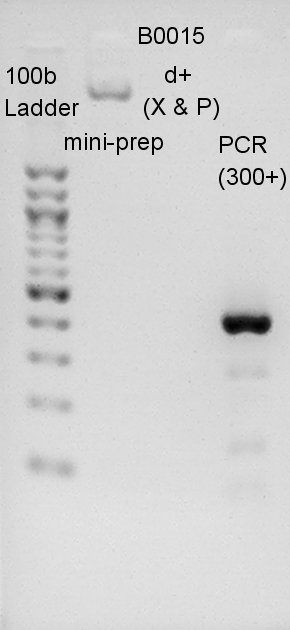
We migrated B0015 mini-prep psoduct, digestion product, and PCR product. PCRed dT would have 429bp(100bp(added by forward primer) + 129bp(dT) + 200bp(added by reverse primer)). Most bright and thick band in this image has about 300~400 bp. We thought dT was amplified successfully.
Gel extraction
Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit(NipponGenetics). Got 50ul of DNA solution.
Digestion
Digestion for dT which amplified with PCR. Digested with XbaI and PstI.
dT
| DNA solution | 5ul |
| XbaI | 1ul |
| PstI | 1ul |
| 10xM buffer | 2ul |
| DW | 11ul |
| Total | 20ul |
Ag43
Digestion result of Ag43 was incorrect. We digested Ag43 once more time.
Digestion
Digestion for Ag43 with SpeI and PstI.
| Ag43 DNA solution | 9ul |
| SpeI | 1ul |
| PstI | 1ul |
| 10xH buffer | 2ul |
| DW | 7ul |
| Total | 20ul |
There are same results with digestion result of recent. We thought PstI would cut different site. What is this 500bp fragment????
Gel extraction
Gel Extraction for digestion products. We used FastGene Gel&PCR extraction kit(NipponGenetics). Got 50ul of DNA solution.
Liquid Culture
Liquid culture for single colony isolated pT7-RBS on pSB1C3 and Ag43-dT on pSB1T3.
Ag43-dT on pSB1T3 colonies were closely existed so we would picked up two or more colonies.
- Picked up one (or tow?) colony from single colony isolated plates by platinum loop.
- Dipped into 2ml of LBC and LBT.
- Cultivated.
</div>
July 17th
digestioned->Ethanol precipitation->Gel Extraction->Ethanol Precipitation
Ag43-dT on pSB1T3 and pT7-RBS on pSB1C3
Mini-prep
Mini-prep for liquid culture products cultivated from yesterday(16th). We used FastGene Plasmid Mini Kit(Nippon Genetics) and got 50ul of DNA solutions.
Electrophoresis
Electrophoresis for digestion product of K346007(Ag43) with EcoRI and SpeI as a reference to confirm SpeI cuts correct site or not and PstI didn't work correctly(in past experiments). And mini-prep products of Ag43-dT on pSB1T3 and pT7-RBS on pSB1C3 also migrated.
If digestion were succeeded, there are two bands would be seen: about 3120bp and 2070bp.
And if ligation-transformation-mini-prep were succeeded, there would be existed two bands in each lanes: about 5000bp and 2000bp.
In this result we confirmed Ag43 was successfully digested with EcoRI and SpeI, and there were no other extra bands which had been existed double digested with EcoRI & PstI and SpeI & PstI. would PstI not work correctly?
About mini-prep products, in Ag43-dT on pSB1T3, there were two plasmid DNA bands about 8000bp and 3000bp in one lane. We thought which means we failed single colony isolation then resuspended two another E.coli colonies another ligated DNA were transformed.
In pT7-RBS on pSB1C3, we needed about 2000bp plasmid DNA and there were about 1500bp of DNA. Plasmid DNA migrates more far than linear DNA so we thought we got correct DNA.
</div>
</div>
</div>
 "
"