Team:Paris Bettencourt/Modeling
From 2012.igem.org
(→Check controls) |
(→Control Checks) |
||
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Paris_Bettencourt/header}} | {{:Team:Paris_Bettencourt/header}} | ||
| - | + | <div id="boston"> | |
| + | <br> | ||
<div id="grouptitle">Safety Assessment</div> | <div id="grouptitle">Safety Assessment</div> | ||
==Overview== | ==Overview== | ||
| - | Safety is an important issue in synthetic biology, especially | + | Safety is an important issue in synthetic biology, especially for environmentally related projects. We started to answer the question, “how safe is safe enough?” by involving experts, the public and our fellow scientists, and also by building biosafety devices. However, to really answer the question, we need first to ask ourselves a more basic question, “how do we measure safety?”. As we see synthetic biology as an engineering approach to biology, we could think about the adaptation of safety engineering, a well studied engineering subset, that has been widely use to minimize risks in many fields of engineering, such as mechanical engineering, aircrafts, and manufactures. However, the risks they face are surely different from the risks of synthetic biology. |
| - | |||
According to Dana et al<sup>[[#References|1]]</sup> there are four areas of risk research in environmental application of synthetic biology: | According to Dana et al<sup>[[#References|1]]</sup> there are four areas of risk research in environmental application of synthetic biology: | ||
*Differences in the physiology of natural and synthetic organisms will affect how they interact with the surrounding environment, | *Differences in the physiology of natural and synthetic organisms will affect how they interact with the surrounding environment, | ||
| Line 13: | Line 13: | ||
*Gene transfer. | *Gene transfer. | ||
| - | Knowing that there are different areas of risk that we need to take into account, we need to design our safety containment with different parts to deal with each of them. Identifying the relationship between one part and the others will help us to see the reliability of the overall system. | + | Knowing that there are different areas of risk that we need to take into account, we need to design our safety containment with different parts to deal with each of them. Identifying the relationship between one part and the others will help us to see the reliability of the overall system. Here we propose an approach to assess safety for environmental release of genetically engineered bacteria (GE bacteria). |
===Objectives=== | ===Objectives=== | ||
| - | *Adapting existing safety assessment tools | + | *Adapting existing safety assessment tools for synthetic biology. |
| - | *Proposing methods to assess safety in synthetic biology. | + | *Proposing new methods to assess safety in synthetic biology. |
==Methods and Results== | ==Methods and Results== | ||
| - | We propose a method to assess hazards and risk<sup>[[#References|2]]</sup> in releasing genetically engineered bacteria | + | We propose a method to assess hazards and risk<sup>[[#References|2]]</sup> in releasing genetically engineered bacteria into the environment. As a case study, we want to release GE bacteria which will perform some function in the environment and we want to apply 3 sets of bWARE safety containment modules (alginate beads, bWARE killswitch, semantic containment) to control the hazards and risks. Here we focus more on the reliability of the safety containment system rather than the functional part, although a similar assessment can also be applied to assess the reliability of the functional part. |
| - | === | + | ===Hazard Identification=== |
| - | Identifying hazards means that we need to find and understand the possible harm that may happen in the application of our system. Generally there are two potential | + | Identifying hazards means that we need to find and understand the possible harm that may happen in the application of our system. Generally there are two potential hazards in environmental application of synthetic biology<sup>[[#References|1]]</sup> which are the successful escape of the GE bacteria followed by successful competition with natural strains, and horizontal gene transfer from GEO to natural strains. |
===Risk Assessment=== | ===Risk Assessment=== | ||
| - | Risk assessment | + | Risk assessment provides an idea of what kind of risk we face in releasing the GEO in the environment and helps to design containment devices. Here we adapted a risk management method in workplaces complying with health and safety law <sup>[[#References|3]]</sup>. |
To-do-list to assess risk in 5 steps: | To-do-list to assess risk in 5 steps: | ||
| Line 47: | Line 47: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| GE bacteria escaping the containment may outcompete natural strains if they have better fitness, creating natural imbalance | | style="border:0.035cm solid #000000;padding:0.176cm;"| GE bacteria escaping the containment may outcompete natural strains if they have better fitness, creating natural imbalance | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| Using harmless strain or strain with low fitness compared to natural strain (standard E coli for lab) | | style="border:0.035cm solid #000000;padding:0.176cm;"| Using harmless strain or strain with low fitness compared to natural strain (standard E coli for lab) | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| Designing a safety containment to prevent the cells reproducing themselves at some point and/or separating them from natural strains | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| Designing a safety containment to prevent the cells from reproducing themselves at some point and/or separating them from natural strains |
|- | |- | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| Horizontal gene transfer (HGT) | | style="border:0.035cm solid #000000;padding:0.176cm;"| Horizontal gene transfer (HGT) | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| Other strain/species may uptake engineered | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| Other strain/species may uptake engineered genes, and if the genes give advantage in fitness, it may outcompete other strains creating natural imbalance |
| style="border:0.035cm solid #000000;padding:0.176cm;"| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| - | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| Designing a safety containment to degrade DNA and/or separate the DNA from the environment and/or prevent natural | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| Designing a safety containment to degrade DNA and/or separate the DNA from the environment and/or prevent natural strains from gaining advantages from modified DNA |
|} | |} | ||
<br> | <br> | ||
| - | === | + | ===Controlling hazards and risks=== |
| - | In this step we decided what control elements we want to | + | In this step we decided what control elements we want to implement in our system to reduce the risks. We should choose the best suitable control elements depending on the system function and application. As we already know that we want to apply bWARE safety containment, here we describe the role of each safety part in risk reduction. |
#GE bacteria outcompete natural strains | #GE bacteria outcompete natural strains | ||
| - | #*In order to prevent | + | #*In order to prevent GE bacteria from escaping and outcompeting the natural strains, we will stop their reproduction by putting a self killing mechanism based on [https://2012.igem.org/Team:Paris_Bettencourt/Suicide suicide] and [https://2012.igem.org/Team:Paris_Bettencourt/Restriction_Enzyme restriction enzyme] systems to kill the cells [https://2012.igem.org/Team:Paris_Bettencourt/Delay after] they perform their function. |
| - | #*In case of the failure of the killing mechanism, we will put the cells inside a physical containment so there will be no physical interaction between them and the surrounding cells. | + | #*In case of the failure of the killing mechanism, we will put the cells inside a [https://2012.igem.org/Team:Paris_Bettencourt/Encapsulation physical containment] so there will be no physical interaction between them and the surrounding cells. |
#HGT | #HGT | ||
| - | #*We will use DNAse to degrade DNA after the cells perform | + | #*We will use a [https://2012.igem.org/Team:Paris_Bettencourt/Suicide DNAse] to degrade DNA after the cells perform their function so they won’t leave any genetic material behind. |
| - | #*In case of the failure | + | #*In case of the failure due to inefficiency of the DNAse, the [https://2012.igem.org/Team:Paris_Bettencourt/Encapsulation physical containment] will still protect the DNA from the environment. |
| - | #*In case of the leakiness of the physical containment, the DNA has special encryption system | + | #*In case of the leakiness of the physical containment, the DNA has a special encryption system, i.e. [https://2012.igem.org/Team:Paris_Bettencourt/Semantic_containment semantic containment], so the receiver cells will not be able to read GE genetic material. |
| - | === | + | ===Control Checks=== |
| - | To check and predict the reliability of our system, we used fault tree analysis to assess multiple safety containment in one biodevice. This top-down approach allows us to see the relationship between each containment element and predict the overall failure probability. With this method we can see what basic events are the key elements in our system. | + | To check and predict the reliability of our system, we used fault tree analysis to assess multiple safety containment modules in one biodevice. This top-down approach allows us to see the relationship between each containment element and predict the overall failure probability. With this method we can see what basic events are the key elements in our system. |
| - | + | [[File:ParisB_FTA.png|800px|center]] | |
AND gate represents independent events | AND gate represents independent events | ||
| Line 108: | Line 108: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| DNA transferred into natural strain | | style="border:0.035cm solid #000000;padding:0.176cm;"| DNA transferred into natural strain | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| HGT | | style="border:0.035cm solid #000000;padding:0.176cm;"| HGT | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| <nowiki>transconjugant to donor ratio in HGT is typically <10</nowiki><sup>-5</sup> <sup>[[#References|4]]</sup> | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| <nowiki>transconjugant to donor ratio in HGT is typically <10</nowiki><sup>-5</sup> <sup>[[#References|4]]</sup>, assume this is the rate per generation |
|- | |- | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| P3 | | style="border:0.035cm solid #000000;padding:0.176cm;"| P3 | ||
| Line 114: | Line 114: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | | style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| HGT | | style="border:0.035cm solid #000000;padding:0.176cm;"| HGT | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation |
|- | |- | ||
| Line 128: | Line 128: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | | style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| no DNA degradation (w/o antitoxin), no death (with antitoxin) | | style="border:0.035cm solid #000000;padding:0.176cm;"| no DNA degradation (w/o antitoxin), no death (with antitoxin) | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation |
|- | |- | ||
| Line 135: | Line 135: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| beneficial mutation | | style="border:0.035cm solid #000000;padding:0.176cm;"| beneficial mutation | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| outcompetition of the mutant | | style="border:0.035cm solid #000000;padding:0.176cm;"| outcompetition of the mutant | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| - |
|- | |- | ||
| Line 142: | Line 142: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | | style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | | style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation |
|- | |- | ||
| Line 149: | Line 149: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | | style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | | style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation |
|- | |- | ||
| Line 156: | Line 156: | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | | style="border:0.035cm solid #000000;padding:0.176cm;"| genetic failure | ||
| style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | | style="border:0.035cm solid #000000;padding:0.176cm;"| no antitoxin degradation | ||
| - | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation | + | | style="border:0.035cm solid #000000;padding:0.176cm;"| estimation |
|} | |} | ||
| - | + | ====Physical containment failure==== | |
| + | |||
| + | From the experiment we get the escape rate of the cells from the alginate beads is (escaping cells/total cells) is 10<sup>-5</sup> per 12 hours. Assuming 30 minutes of division time, we have 24 generations, and the escape rate per generation is 4 x 10<sup>-6</sup>. | ||
| + | |||
| + | ====Genetic failure==== | ||
There are different processes that lead to a loss of function of a gene | There are different processes that lead to a loss of function of a gene | ||
| Line 172: | Line 176: | ||
Estimating the mutation rate which cause failure of the safety containment of the device | Estimating the mutation rate which cause failure of the safety containment of the device | ||
| - | * Spontaneous mutation rate of a wild-type E coli strain growing on research medium is 3. | + | * Spontaneous mutation rate of a wild-type E coli strain growing on research medium is 3.3 x 10<sup>-9</sup> per nucleotide per generation <sup>[[#References|5]]</sup> |
* approximately ⅔ of nucleotide mutation will change amino acid (64 combination of nucleotides code only 20 amino acids) | * approximately ⅔ of nucleotide mutation will change amino acid (64 combination of nucleotides code only 20 amino acids) | ||
* approximately 70% of amino acid change will lose the part function <sup>[[#References|6]]</sup> | * approximately 70% of amino acid change will lose the part function <sup>[[#References|6]]</sup> | ||
| Line 180: | Line 184: | ||
P<sub>L</sub><nowiki>= 3.3 x 10</nowiki><sup>-9</sup> x ⅔ x 0.70 x 1000 = 1.54 x 10<sup>-6</sup> | P<sub>L</sub><nowiki>= 3.3 x 10</nowiki><sup>-9</sup> x ⅔ x 0.70 x 1000 = 1.54 x 10<sup>-6</sup> | ||
| - | |||
| - | |||
'''Recombination''' | '''Recombination''' | ||
| + | Finding a definite rate of recombination is not easy. Guttman and Dykhuizen in 1994 <sup>[[#References|8]]</sup> stated that recombination is believed to occur at such a low frequency, but their experiment showed that the rate could be as high as mutation rate (50 fold higher compared to the mutation rate believed at that moment, but actually it is 5.0 x 10<sup>-9</sup> changes per nucleotide per generation, a bit more than one fold to the value believed now). | ||
| + | |||
| + | Redoing the previous calculation with this number will lead us to a rate of 2.33 x 10<sup>-6</sup> per gene per generation. | ||
'''Plasmid loss''' | '''Plasmid loss''' | ||
| Line 190: | Line 195: | ||
According to L Boe <sup>[[#References|9]]</sup> plasmid loss rate is defined as the probability that a division of a plasmid-carrying individual results in the birth of one plasmid-free and one plasmid-carrying daughter cell. For the low copy plasmid this rate is about ~1% /division and for high copy plasmid is ~0.01% /division, neglecting the viable/non-viable factor of the cells of losing the plasmid (no selection). If a plasmid with a lost rate of 0.05 is stabilized with a 100% efficient postsegragational killing system, the 5% of the division will result in non viable daughter cell, and we can not see the effect except of increasing the appearing division time by ln 2/ln (1-0.05) = 1.04. Thus in our system we can neglect this plasmid loss assuming that we have antibiotic selection in the beads. | According to L Boe <sup>[[#References|9]]</sup> plasmid loss rate is defined as the probability that a division of a plasmid-carrying individual results in the birth of one plasmid-free and one plasmid-carrying daughter cell. For the low copy plasmid this rate is about ~1% /division and for high copy plasmid is ~0.01% /division, neglecting the viable/non-viable factor of the cells of losing the plasmid (no selection). If a plasmid with a lost rate of 0.05 is stabilized with a 100% efficient postsegragational killing system, the 5% of the division will result in non viable daughter cell, and we can not see the effect except of increasing the appearing division time by ln 2/ln (1-0.05) = 1.04. Thus in our system we can neglect this plasmid loss assuming that we have antibiotic selection in the beads. | ||
| - | + | Assuming only mutation and recombination could happen to delete a function of a gene, we have the total genetic failure rate approximately 4 x 10<sup>-6</sup> per gene per generation. | |
| - | + | ====Total failure probability==== | |
| + | |||
| + | Having Ptotal = P1.P2.P3.P4.P5 + P1.P6.(P5+P7+P8+P9) and put 1 for unknown probability (P4 and P6) as a worst-case scenario, we come to the failure rate number 6.4 x 10<sup>-11</sup> per generation. | ||
| + | |||
| + | A volume of one alginate bead is approximately 20 uL. So if the cells grow until they reach the stationary phase (4 x 10<sup>9</sup> cells/ml), we will have 8 x 10<sup>7</sup> cells. If we started from 100 cells per bead, the number of generations we have is <sup>2</sup>log (8 x 10<sup>7</sup> / 100) =19.6 generations. Therefore the total failure becomes 6.4 x 10<sup>-11</sup> x 19.6 = 1.25 x 10<sup>-9</sup>. | ||
| + | |||
| + | With this method we can also see that the sense-HGT rate is much lower (6.4 x 10<sup>-22</sup>) , almost negligible compared to the outcompetition rate, because it has more back up/redundancy components. | ||
== Discussion == | == Discussion == | ||
| - | |||
| + | We have proposed and implemented an example of assessing biosafety by adapting existing methods from safety engineering. However, it is very clear that the prediction is based on rough estimation and we need more experimental data to verify this estimation. For an example, Torres et al <sup>[[#References|10]]</sup> tested a dual containment killing system and compared it with single containment system. They tested an EcoRI based system and a Colicin E3-based system and obtained the efficacy (based on the numbers of successful transformants) each 10<sup>4</sup> and 10<sup>5</sup> respectively but when they combine both system they only got efficacy 10<sup>6</sup>, while theoretically they could reach up to 10<sup>9</sup>. This could give us an idea that predicting the performance of a combination of genetic parts is not as easy as combining the performance of each genetic part. However, we can still conclude that having redundant parts is necessary to reduce the failure rate and make it as low as possible. | ||
| + | |||
| + | Research in measuring the evolutionary stability in E coli was performed by Sean C Sleight et al <sup>[[#References|11]]</sup> where they measured it of BioBrick-assembled genetic circuits in E coli over multiple generation by measuring the number of loss-functioning mutants. They concluded that to make a robust GE bacteria, one has to take into account 3 principles: | ||
# High expression of genetic circuits comes with the cost of low evolutionary stability (for example induced vs non-induced system) | # High expression of genetic circuits comes with the cost of low evolutionary stability (for example induced vs non-induced system) | ||
| - | |||
#Avoid repeated sequences because it will more likely be mutated | #Avoid repeated sequences because it will more likely be mutated | ||
| - | |||
#Use of inducible promoters generally increases evolutionary stability compared to constitutive promoters | #Use of inducible promoters generally increases evolutionary stability compared to constitutive promoters | ||
| - | To predict the robustness of the system from mutation, we have to take into account | + | To predict the robustness of the system from mutation, we have to take into account the duration (i.e. how many generations) we want our system to work. From their experiment for example, the AHL induced TetR-GFP cells lose their functions after 30 generations, while the uninduced ones lose their function slower (50% after 300 generations). A similar idea is also shown from our work where the mutation rate we got from literature research is in the unit of per generation. This is how reproduction differs biological system from other engineering systems. In engineering we also have failure probability as a function of time because of the life cycle of the system, but the system is not reproducing so it has no variability. |
| - | We need a method to predict the evolutionary stability of circuits from the properties of their parts, but the emergent behaviours of circuits will likely make prediction difficult. Thus it will be very useful if each BioBricks parts is completed with evolutionary stability data sheet, so we can make prediction of the stability of more complex circuits. | + | Analyzing key elements that cause failure in any system including synthetic biology systems is essential for system improvement and prediction of failure. Adaptation of classical safety engineering methods needs to take into account the unique properties of synthetic biology. Reproducibility and complexity are two examples of areas that need to be studied deeper in this context. We invite future iGEM teams to use and improve our assessment framework, build a solid assessment protocol towards a better understanding of biosafety and successful application of synthetic biology. |
| + | |||
| + | == Conclusion == | ||
| + | *We can adapt methods in safety engineering to assess risk in biosafety | ||
| + | *We need more experimental data to confirm the risk assessing method | ||
| + | |||
| + | == Perspectives == | ||
| + | |||
| + | *We need a method to predict the evolutionary stability of circuits from the properties of their parts, but the emergent behaviours of circuits will likely make prediction difficult. Thus it will be very useful if each BioBricks parts is completed with evolutionary stability data sheet, so we can make prediction of the stability of more complex circuits. | ||
| + | *This safety assessment method is useful for identifying key elements of having failure in synthetic circuit systems before we go further to the real application. Thus we recommend for the future iGEM team to use our assessment method and give feedback to the community by improving this framework. | ||
==References== | ==References== | ||
| Line 214: | Line 234: | ||
[[#Methods_and_Results|3]] - “Five steps to risk assessment” from [http://www.hse.gov.uk/risk/fivesteps.htm http://www.hse.gov.uk/risk/fivesteps.htm]. - risk management method in workplaces complying with health and safety law | [[#Methods_and_Results|3]] - “Five steps to risk assessment” from [http://www.hse.gov.uk/risk/fivesteps.htm http://www.hse.gov.uk/risk/fivesteps.htm]. - risk management method in workplaces complying with health and safety law | ||
| - | [[# | + | [[#Check_controls|4]] - Soren J Sorensen et al. 2005. Studying plasmid horizontal gene transfer in situ: a critical review. Nature Reviews Microbiology Vol 3 |
| + | |||
| + | [[#Check_controls|5]] - Heewook Lee et al., Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. 2012. PNAS E2774-E2783 | ||
| + | |||
| + | [[#Check_controls|6]] - Stanley A. Sawyer et al. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. 2007. PNAS 104(16):6504-6510 | ||
| - | [[# | + | [[#Check_controls|7]] - Lin Xu et al. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between two kingdoms. 2006. Mol. Biol. Evol. 23(6):1107-1108 |
| - | [[# | + | [[#Check_controls|8]] - David S Guttmand and Daniel E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science Vol 266 |
| - | [[# | + | [[#Check_controls|9]] - L.Boe. 1996. Estimation of Plasmid Loss Rates in Bacterial Populations with a Reference to the Reproducibility of Stability Experiments. Plasmid 36, 161–167. Article No. 0043 |
| - | [[# | + | [[#Discussion|10]] - Torres B et al. 2003. A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology. 2003 Dec;149(Pt 12):3595-601. |
| + | [[#Discussion|11]] - Sean C Sleight et al., Designing and engineering evolutionary robust genetic circuits. 2010. Journal of Biological Engineering, 4:12 | ||
| + | </div> | ||
{{:Team:Paris_Bettencourt/footer}} | {{:Team:Paris_Bettencourt/footer}} | ||
Latest revision as of 02:47, 27 October 2012
Contents |
Overview
Safety is an important issue in synthetic biology, especially for environmentally related projects. We started to answer the question, “how safe is safe enough?” by involving experts, the public and our fellow scientists, and also by building biosafety devices. However, to really answer the question, we need first to ask ourselves a more basic question, “how do we measure safety?”. As we see synthetic biology as an engineering approach to biology, we could think about the adaptation of safety engineering, a well studied engineering subset, that has been widely use to minimize risks in many fields of engineering, such as mechanical engineering, aircrafts, and manufactures. However, the risks they face are surely different from the risks of synthetic biology.
According to Dana et al1 there are four areas of risk research in environmental application of synthetic biology:
- Differences in the physiology of natural and synthetic organisms will affect how they interact with the surrounding environment,
- Escaped microorganisms have the potential to survive in receiving environments and to compete successfully with non-modified counterparts,
- Synthetic organisms might evolve and adapt quickly, perhaps filling new ecological niches, and
- Gene transfer.
Knowing that there are different areas of risk that we need to take into account, we need to design our safety containment with different parts to deal with each of them. Identifying the relationship between one part and the others will help us to see the reliability of the overall system. Here we propose an approach to assess safety for environmental release of genetically engineered bacteria (GE bacteria).
Objectives
- Adapting existing safety assessment tools for synthetic biology.
- Proposing new methods to assess safety in synthetic biology.
Methods and Results
We propose a method to assess hazards and risk2 in releasing genetically engineered bacteria into the environment. As a case study, we want to release GE bacteria which will perform some function in the environment and we want to apply 3 sets of bWARE safety containment modules (alginate beads, bWARE killswitch, semantic containment) to control the hazards and risks. Here we focus more on the reliability of the safety containment system rather than the functional part, although a similar assessment can also be applied to assess the reliability of the functional part.
Hazard Identification
Identifying hazards means that we need to find and understand the possible harm that may happen in the application of our system. Generally there are two potential hazards in environmental application of synthetic biology1 which are the successful escape of the GE bacteria followed by successful competition with natural strains, and horizontal gene transfer from GEO to natural strains.
Risk Assessment
Risk assessment provides an idea of what kind of risk we face in releasing the GEO in the environment and helps to design containment devices. Here we adapted a risk management method in workplaces complying with health and safety law 3.
To-do-list to assess risk in 5 steps:
- Identify the hazards
- Decide who might be harmed and how
- Evaluate the risks and decide on precaution
- Record your findings and implement them
- Review your assessment and update if necessary
Worksheet example:
| What are the hazards? | Who might be harmed and how? | What are we already doing? | What further action is necessary? |
| GE bacteria outcompete natural strains | GE bacteria escaping the containment may outcompete natural strains if they have better fitness, creating natural imbalance | Using harmless strain or strain with low fitness compared to natural strain (standard E coli for lab) | Designing a safety containment to prevent the cells from reproducing themselves at some point and/or separating them from natural strains |
| Horizontal gene transfer (HGT) | Other strain/species may uptake engineered genes, and if the genes give advantage in fitness, it may outcompete other strains creating natural imbalance | - | Designing a safety containment to degrade DNA and/or separate the DNA from the environment and/or prevent natural strains from gaining advantages from modified DNA |
Controlling hazards and risks
In this step we decided what control elements we want to implement in our system to reduce the risks. We should choose the best suitable control elements depending on the system function and application. As we already know that we want to apply bWARE safety containment, here we describe the role of each safety part in risk reduction.
- GE bacteria outcompete natural strains
- In order to prevent GE bacteria from escaping and outcompeting the natural strains, we will stop their reproduction by putting a self killing mechanism based on suicide and restriction enzyme systems to kill the cells after they perform their function.
- In case of the failure of the killing mechanism, we will put the cells inside a physical containment so there will be no physical interaction between them and the surrounding cells.
- HGT
- We will use a DNAse to degrade DNA after the cells perform their function so they won’t leave any genetic material behind.
- In case of the failure due to inefficiency of the DNAse, the physical containment will still protect the DNA from the environment.
- In case of the leakiness of the physical containment, the DNA has a special encryption system, i.e. semantic containment, so the receiver cells will not be able to read GE genetic material.
Control Checks
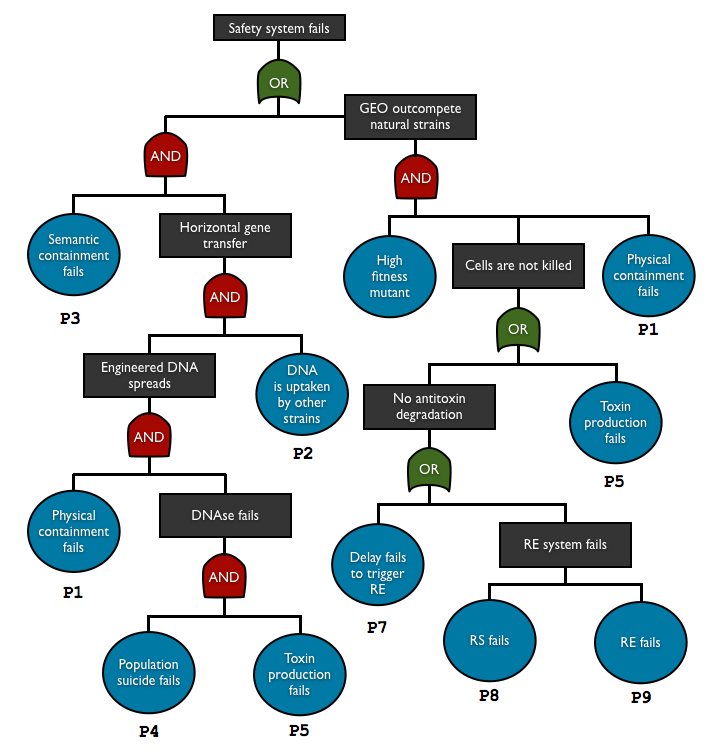
To check and predict the reliability of our system, we used fault tree analysis to assess multiple safety containment modules in one biodevice. This top-down approach allows us to see the relationship between each containment element and predict the overall failure probability. With this method we can see what basic events are the key elements in our system.
AND gate represents independent events
P(A and B) = P(A)P(B)
OR gate corresponds to set of union
P(A or B) = P(A) + P(B)
so the total failure probability of the system is
Ptotal = P1.P2.P3.P4.P5 + P1.P6.(P5+P7+P8+P9)
List of the basic events
| Notation | Failure component | Failure mode | Consequence | Method for determining the probability |
| P1 | physical containment | leakage | DNA/cell escape | experiment |
| P2 | DNA uptake | DNA transferred into natural strain | HGT | transconjugant to donor ratio in HGT is typically <10-5 4, assume this is the rate per generation |
| P3 | semantic containment | genetic failure | HGT | estimation |
| P4 | population suicide | no surrounding cells with enough toxin | no death | based on the cell density and the expectation number of surrounding non-mutant |
| P5 | toxin production | genetic failure | no DNA degradation (w/o antitoxin), no death (with antitoxin) | estimation |
| P6 | mutant fitness | beneficial mutation | outcompetition of the mutant | - |
| P7 | delay system | genetic failure | no antitoxin degradation | estimation |
| P8 | restriction enzyme | genetic failure | no antitoxin degradation | estimation |
| P9 | restriction site | genetic failure | no antitoxin degradation | estimation |
Physical containment failure
From the experiment we get the escape rate of the cells from the alginate beads is (escaping cells/total cells) is 10-5 per 12 hours. Assuming 30 minutes of division time, we have 24 generations, and the escape rate per generation is 4 x 10-6.
Genetic failure
There are different processes that lead to a loss of function of a gene
- Mutation
- Recombination
- Plasmid loss
Losing function because of mutation
Estimating the mutation rate which cause failure of the safety containment of the device
- Spontaneous mutation rate of a wild-type E coli strain growing on research medium is 3.3 x 10-9 per nucleotide per generation 5
- approximately ⅔ of nucleotide mutation will change amino acid (64 combination of nucleotides code only 20 amino acids)
- approximately 70% of amino acid change will lose the part function 6
- mean length of genetic coding sequence for E coli (procaryotes) is 924 bp 7 ≈ 1000 bp
So the loss function rate of a gene per generation because of mutation is
PL= 3.3 x 10-9 x ⅔ x 0.70 x 1000 = 1.54 x 10-6
Recombination
Finding a definite rate of recombination is not easy. Guttman and Dykhuizen in 1994 8 stated that recombination is believed to occur at such a low frequency, but their experiment showed that the rate could be as high as mutation rate (50 fold higher compared to the mutation rate believed at that moment, but actually it is 5.0 x 10-9 changes per nucleotide per generation, a bit more than one fold to the value believed now).
Redoing the previous calculation with this number will lead us to a rate of 2.33 x 10-6 per gene per generation.
Plasmid loss
According to L Boe 9 plasmid loss rate is defined as the probability that a division of a plasmid-carrying individual results in the birth of one plasmid-free and one plasmid-carrying daughter cell. For the low copy plasmid this rate is about ~1% /division and for high copy plasmid is ~0.01% /division, neglecting the viable/non-viable factor of the cells of losing the plasmid (no selection). If a plasmid with a lost rate of 0.05 is stabilized with a 100% efficient postsegragational killing system, the 5% of the division will result in non viable daughter cell, and we can not see the effect except of increasing the appearing division time by ln 2/ln (1-0.05) = 1.04. Thus in our system we can neglect this plasmid loss assuming that we have antibiotic selection in the beads.
Assuming only mutation and recombination could happen to delete a function of a gene, we have the total genetic failure rate approximately 4 x 10-6 per gene per generation.
Total failure probability
Having Ptotal = P1.P2.P3.P4.P5 + P1.P6.(P5+P7+P8+P9) and put 1 for unknown probability (P4 and P6) as a worst-case scenario, we come to the failure rate number 6.4 x 10-11 per generation.
A volume of one alginate bead is approximately 20 uL. So if the cells grow until they reach the stationary phase (4 x 109 cells/ml), we will have 8 x 107 cells. If we started from 100 cells per bead, the number of generations we have is 2log (8 x 107 / 100) =19.6 generations. Therefore the total failure becomes 6.4 x 10-11 x 19.6 = 1.25 x 10-9.
With this method we can also see that the sense-HGT rate is much lower (6.4 x 10-22) , almost negligible compared to the outcompetition rate, because it has more back up/redundancy components.
Discussion
We have proposed and implemented an example of assessing biosafety by adapting existing methods from safety engineering. However, it is very clear that the prediction is based on rough estimation and we need more experimental data to verify this estimation. For an example, Torres et al 10 tested a dual containment killing system and compared it with single containment system. They tested an EcoRI based system and a Colicin E3-based system and obtained the efficacy (based on the numbers of successful transformants) each 104 and 105 respectively but when they combine both system they only got efficacy 106, while theoretically they could reach up to 109. This could give us an idea that predicting the performance of a combination of genetic parts is not as easy as combining the performance of each genetic part. However, we can still conclude that having redundant parts is necessary to reduce the failure rate and make it as low as possible.
Research in measuring the evolutionary stability in E coli was performed by Sean C Sleight et al 11 where they measured it of BioBrick-assembled genetic circuits in E coli over multiple generation by measuring the number of loss-functioning mutants. They concluded that to make a robust GE bacteria, one has to take into account 3 principles:
- High expression of genetic circuits comes with the cost of low evolutionary stability (for example induced vs non-induced system)
- Avoid repeated sequences because it will more likely be mutated
- Use of inducible promoters generally increases evolutionary stability compared to constitutive promoters
To predict the robustness of the system from mutation, we have to take into account the duration (i.e. how many generations) we want our system to work. From their experiment for example, the AHL induced TetR-GFP cells lose their functions after 30 generations, while the uninduced ones lose their function slower (50% after 300 generations). A similar idea is also shown from our work where the mutation rate we got from literature research is in the unit of per generation. This is how reproduction differs biological system from other engineering systems. In engineering we also have failure probability as a function of time because of the life cycle of the system, but the system is not reproducing so it has no variability.
Analyzing key elements that cause failure in any system including synthetic biology systems is essential for system improvement and prediction of failure. Adaptation of classical safety engineering methods needs to take into account the unique properties of synthetic biology. Reproducibility and complexity are two examples of areas that need to be studied deeper in this context. We invite future iGEM teams to use and improve our assessment framework, build a solid assessment protocol towards a better understanding of biosafety and successful application of synthetic biology.
Conclusion
- We can adapt methods in safety engineering to assess risk in biosafety
- We need more experimental data to confirm the risk assessing method
Perspectives
- We need a method to predict the evolutionary stability of circuits from the properties of their parts, but the emergent behaviours of circuits will likely make prediction difficult. Thus it will be very useful if each BioBricks parts is completed with evolutionary stability data sheet, so we can make prediction of the stability of more complex circuits.
- This safety assessment method is useful for identifying key elements of having failure in synthetic circuit systems before we go further to the real application. Thus we recommend for the future iGEM team to use our assessment method and give feedback to the community by improving this framework.
References
1 - Genya V Dana et al. Four steps to avoid a synthetic biology disaster. 2012. Nature vol 483
2 - WorkSafe Victoria. A handbook for workplaces, controlling OHS hazards and risks. 2007. Edition No.1
3 - “Five steps to risk assessment” from [http://www.hse.gov.uk/risk/fivesteps.htm http://www.hse.gov.uk/risk/fivesteps.htm]. - risk management method in workplaces complying with health and safety law
4 - Soren J Sorensen et al. 2005. Studying plasmid horizontal gene transfer in situ: a critical review. Nature Reviews Microbiology Vol 3
5 - Heewook Lee et al., Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. 2012. PNAS E2774-E2783
6 - Stanley A. Sawyer et al. Prevalence of positive selection among nearly neutral amino acid replacements in Drosophila. 2007. PNAS 104(16):6504-6510
7 - Lin Xu et al. Average gene length is highly conserved in prokaryotes and eukaryotes and diverges only between two kingdoms. 2006. Mol. Biol. Evol. 23(6):1107-1108
8 - David S Guttmand and Daniel E. Dykhuizen. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science Vol 266
9 - L.Boe. 1996. Estimation of Plasmid Loss Rates in Bacterial Populations with a Reference to the Reproducibility of Stability Experiments. Plasmid 36, 161–167. Article No. 0043
10 - Torres B et al. 2003. A dual lethal system to enhance containment of recombinant micro-organisms. Microbiology. 2003 Dec;149(Pt 12):3595-601.
11 - Sean C Sleight et al., Designing and engineering evolutionary robust genetic circuits. 2010. Journal of Biological Engineering, 4:12
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment