Team:HokkaidoU Japan/Notebook/aggregation Week 12
From 2012.igem.org
(17~20th images) |
|||
| (13 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | <!-- DO NOT EDIT OVER THIS LINE @iTakeshi --> | ||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 17th== | + | ===September 17th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Digestion of RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 and ptet-pSB1A2== | + | ====Digestion of RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 and ptet-pSB1A2==== |
| - | + | To make a construct of ptet-RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 with XbaI and PstI, ptet-pSB1A2 with SpeI and PstI. | |
| - | To make a construct of ptet-RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 with XbaI | + | <br /> |
| - | + | ||
Insert (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2) | Insert (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2) | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| - | |DNA solution ( 45 | + | |DNA solution (45~50 ng/ul) |
|27 ul | |27 ul | ||
|- | |- | ||
| Line 96: | Line 95: | ||
| - | + | ====Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 15 min at 4C. |
| - | # | + | #Removed supernatant and added 220 ul of 70% ethanol. |
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 10 min at 4C. |
| - | # | + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. |
| - | + | ||
| - | |||
We confirmed that the concentration of Insert DNA solution is 60~70 ng/ul and Vector DNA solution is about 20~30 ng/ul. | We confirmed that the concentration of Insert DNA solution is 60~70 ng/ul and Vector DNA solution is about 20~30 ng/ul. | ||
| - | |||
| - | ==Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | ====Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 148: | Line 138: | ||
|} | |} | ||
| - | + | ====Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2==== | |
| - | + | Transformation of ligation ligation product into JM109. | |
| - | ==Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2== | + | |
| - | + | ||
| - | Transformation of JM109. | + | |
#Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | ||
#Incubated on ice for 30 min. | #Incubated on ice for 30 min. | ||
| Line 159: | Line 146: | ||
#Plated 300 ul of the culture onto first dish and spread. | #Plated 300 ul of the culture onto first dish and spread. | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| - | #Incubated the plates at 37C for 17 | + | #Incubated the plates at 37C for 17 hrs. |
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | |||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 18th== | + | ===September 18th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3== | + | ====Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3==== |
| - | + | ||
We did colony PCR two times. | We did colony PCR two times. | ||
| - | |||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 225: | Line 211: | ||
|} | |} | ||
Cycle:2~4 x 35 | Cycle:2~4 x 35 | ||
| - | + | ||
We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls. | We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls. | ||
| Line 233: | Line 219: | ||
[[image:HokkaidoU2012 120918 colop2 pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3.jpg|thumb|Colony PCR result 2]] | [[image:HokkaidoU2012 120918 colop2 pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3.jpg|thumb|Colony PCR result 2]] | ||
| + | Target products didn't exist in all samples. We noticed the reason why such results shown is that we used incorrect pSB1C3 DNA solution which isn't confirmed the sequence. We'll try the synthesis once more. | ||
| - | + | ====Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pSB1C3==== | |
| - | + | To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI and SpeI and pSB1C3 with EcoRI and SpeI. | |
| - | + | <br /> | |
| - | ==Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pSB1C3== | + | |
| - | + | ||
| - | To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI | + | |
| - | + | ||
Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28) | Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28) | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
| Line 304: | Line 287: | ||
| - | [[image:|thumb|digestion result]] | + | [[image:HokkaidoU2012 120916 Digestion Insert Vector Control.jpg |thumb|digestion result]] |
| - | + | ====Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3==== | |
| - | ==Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3== | + | |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 15 min at 4C. |
| - | # | + | #Removed supernatant and added 220 ul of 70% ethanol. |
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 10 min at 4C. |
| - | # | + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. |
| + | [[image:HokkaidoU2012 120916 Ethapre Insert Vector.jpg|thumb|ethanol precipitation result]] | ||
| - | + | ====Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3==== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | ==Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3== | + | |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 355: | Line 332: | ||
|} | |} | ||
| - | + | ====Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3==== | |
| - | + | Transformation ligation product into JM109. | |
| - | ==Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3== | + | |
| - | + | ||
| - | Transformation | + | |
#Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice. | ||
#Incubated on ice for 30 min. | #Incubated on ice for 30 min. | ||
| Line 367: | Line 341: | ||
#Plated 300 ul of the culture onto first dish and spread. | #Plated 300 ul of the culture onto first dish and spread. | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| - | #Incubated the plates at 37C for 15 | + | #Incubated the plates at 37C for 15 hrs. |
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 19th== | + | ===September 19th=== |
| - | <div> | + | <div class="hokkaidou-section"> |
| - | ==Colony PCR of pBAD-RBS-eCFP-RBS-Ag43- | + | ====Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28==== |
| - | + | ||
| - | + | ||
Colony PCR to confirm whether pSTV28 vettor has pBAS-RBs-eCFP-RBs-Ag43-dT as insert or not. | Colony PCR to confirm whether pSTV28 vettor has pBAS-RBs-eCFP-RBs-Ag43-dT as insert or not. | ||
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
| Line 432: | Line 404: | ||
|} | |} | ||
Cycle:2~4 x 35 | Cycle:2~4 x 35 | ||
| - | + | ||
We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls. | We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls. | ||
Desired product is about 702bp. | Desired product is about 702bp. | ||
| - | [[image:|thumb|Colony PCR result]] | + | [[image:HokkaidoU2012 120919 colop2 pBAD-RBS-eCFP-RBS-AG43-dT-pSTV28(pSB1C3).jpg|thumb|Colony PCR result]] |
We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA. | We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA. | ||
| - | |||
| - | ==Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3 and pBAD-RBS-eCFP-RBS-Ag43-dI | + | ====Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3 and pBAD-RBS-eCFP-RBS-Ag43-dI on pSB1C3==== |
| - | + | ||
| - | Colony PCR to confirm whether the constructs written above really have eCFP and Ag43 coding sites or not by using primers: one can | + | Colony PCR to confirm whether the constructs written above really have eCFP and Ag43 coding sites or not by using primers: one can anneal to a part of eCFP coding site and another can anneal to a part of Ag43 coding site. |
{|class="hokkaidou-table-pcr-reagent" | {|class="hokkaidou-table-pcr-reagent" | ||
|- | |- | ||
| Line 499: | Line 469: | ||
|} | |} | ||
Cycle:2~4 x 35 | Cycle:2~4 x 35 | ||
| - | |||
We used N1, N2 (DW only) as controls. | We used N1, N2 (DW only) as controls. | ||
Desired product is about 452 bp and 697 bp respectively. | Desired product is about 452 bp and 697 bp respectively. | ||
| - | [[image:|thumb|Colony PCR result]] | + | [[image:HokkaidoU2012 120919 colop3 pBAD-RBS-eCFP-RBS-AG43-dT-pSTV28(pSB1C3).jpg|thumb|Colony PCR result]] |
We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA. | We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA. | ||
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
| - | |||
| - | |||
<div class="hokkaidou-notebook-daily"> | <div class="hokkaidou-notebook-daily"> | ||
| - | ==September 20th== | + | ===September 20th=== |
| - | <div | + | <div class="hokkaidou-section"> |
| - | + | ||
| - | + | ||
| - | + | ||
| + | ====Digestion of ptet-RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 (DNA solution eluted from colony No.10, 12 respectively in ) and ptet-pSB1A2==== | ||
| + | To make a construct of ptet-RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 with XbaI and PstI, ptet-pSB1A2 with SpeI and PstI. We prepared DNa solution derived from No. 10 and No. 12 colony selected selected by the result of colony PCR for 18th. | ||
| + | <br /> | ||
Insert No.10 (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2) | Insert No.10 (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2) | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| - | |DNA solution ( 40 ng/ul) | + | |DNA solution (40 ng/ul) |
|20 ul | |20 ul | ||
|- | |- | ||
| Line 545: | Line 513: | ||
{|class="hokkaidou-table-digestion" | {|class="hokkaidou-table-digestion" | ||
|- | |- | ||
| - | |DNA solution ( 40 ng/ul) | + | |DNA solution (40 ng/ul) |
|20 ul | |20 ul | ||
|- | |- | ||
| Line 610: | Line 578: | ||
[[image:HokkaidoU2012 120919 digestion InsertNo10-InsertNo12-Vector.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120919 digestion InsertNo10-InsertNo12-Vector.jpg|thumb|digestion result]] | ||
| - | + | ====Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== | |
| - | + | ||
| - | ==Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | |
| - | + | ||
#Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | #Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol. | ||
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 15 min at 4C. |
| - | # | + | #Removed supernatant and added 220 ul of 70% ethanol. |
| - | #Centrifuged | + | #Centrifuged at 15000 rpm, 10 min at 4C. |
| - | # | + | #Removed supernatant and dried out at room temperature, after that added 10 ul of DW. |
| - | [[image:|thumb|ethanol precipitation result]] | + | [[image:HokkaidoU2012 120919 Ethapre InsertNo10-InsertNo12-Vector.jpg|thumb|ethanol precipitation result]] |
| - | We decided to use No.10 digestion product for ligation, and confirmed that the concentration of Insert DNA solution is 50 ng/ul and Vector DNA solution is about 20~30 ng/ul. | + | We decided to use No. 10 digestion product for ligation, and confirmed that the concentration of Insert DNA solution is 50 ng/ul and Vector DNA solution is about 20~30 ng/ul. |
| - | + | ||
| - | ==Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2== | + | ====Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2==== |
| - | + | ||
{|class="hokkaidou-table-ligation" | {|class="hokkaidou-table-ligation" | ||
|- | |- | ||
| Line 663: | Line 626: | ||
|} | |} | ||
| - | |||
| - | ==Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2== | + | ====Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2==== |
| - | + | Transformation of ligation product into JM109. | |
| - | Transformation of JM109. | + | |
#Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | #Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice. | ||
#Incubated on ice for 30 min. | #Incubated on ice for 30 min. | ||
| Line 675: | Line 636: | ||
#Plated 300 ul of the culture onto first dish and spread. | #Plated 300 ul of the culture onto first dish and spread. | ||
#Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | #Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread. | ||
| - | #Incubated the plates at 37C for 15 | + | #Incubated the plates at 37C for 15 hrs. |
| - | </ | + | |
| + | <br style="line-height: 0; clear: both;" /> | ||
| + | |||
| + | </div></div> | ||
| + | |||
| + | <div class="hokkaidou-notebook-daily"> | ||
| + | ===September 21th=== | ||
| + | <div class="hokkaidou-section"> | ||
| + | ====Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3==== | ||
| + | |||
| + | {|class="hokkaidou-table-pcr-reagent" | ||
| + | |- | ||
| + | |DNA solution | ||
| + | |4 ul | ||
| + | |- | ||
| + | |Kapa-Taq(Taq polymerase) | ||
| + | |10 ul | ||
| + | |- | ||
| + | |Forward Primer(X-phaB-F primer) | ||
| + | |0.8 ul | ||
| + | |- | ||
| + | |Reverse Primer(PS-R primer) | ||
| + | |0.8 ul | ||
| + | |- | ||
| + | |DW | ||
| + | |4.4 ul | ||
| + | |- | ||
| + | |Total | ||
| + | |20 ul | ||
| + | |} | ||
| + | |||
| + | |||
| + | {|class="hokkaidou-table-pcr-time" | ||
| + | |- | ||
| + | |Number | ||
| + | |Degree | ||
| + | |Second | ||
| + | |- | ||
| + | |1 | ||
| + | |95 | ||
| + | |120 | ||
| + | |- | ||
| + | |2 | ||
| + | |95 | ||
| + | |30 | ||
| + | |- | ||
| + | |3 | ||
| + | |68.9 | ||
| + | |30 | ||
| + | |- | ||
| + | |4 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |5 | ||
| + | |72 | ||
| + | |60 | ||
| + | |- | ||
| + | |6 | ||
| + | |4 | ||
| + | |HOLD | ||
| + | |} | ||
| + | Cycle:2~4 x 35 | ||
| + | |||
| + | |||
| + | We used N1 (DW only) and N2(RBS-phaC-RBs-phaA-RBS-phaB-pSB1A2)as controls. | ||
| + | Desired product is about 1500bp. | ||
| + | |||
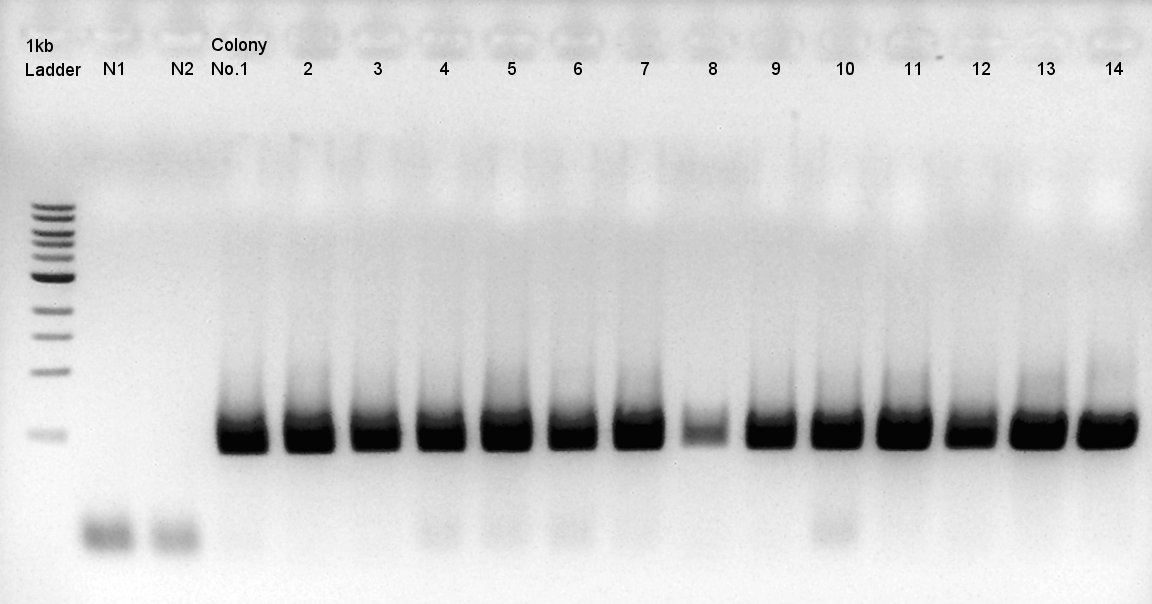
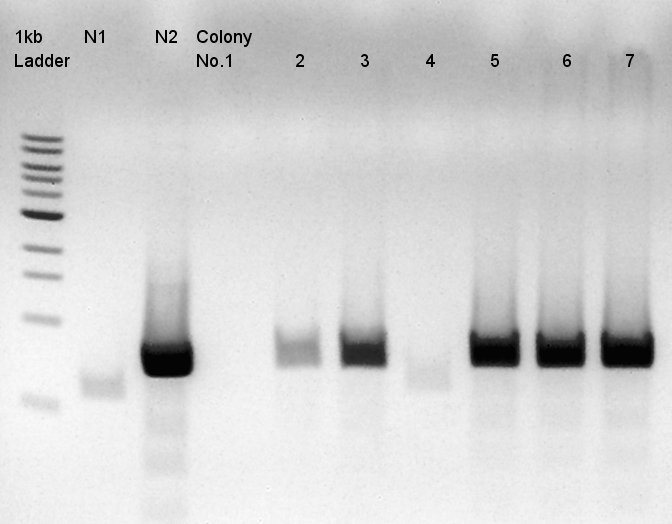
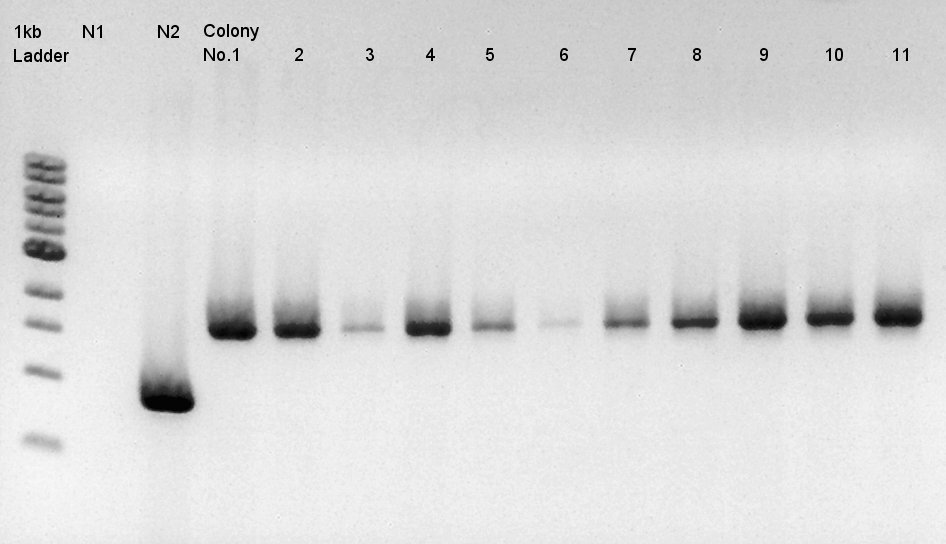
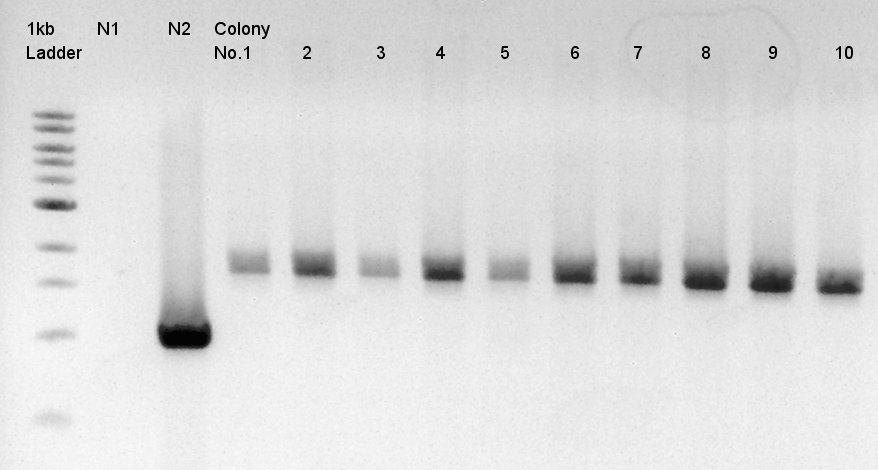
| + | [[image:HokkaidoU2012 120921 colop1 ptet-RBS-phaCAB-RBS-eYFP-dT-pSB1C3.jpg|thumb|Colony PCR result]] | ||
| + | [[image:HokkaidoU2012 120921 colop2 ptet-RBS-phaCAB-RBS-eYFP-dT-pSB1C3.jpg|thumb|Colony PCR result2]] | ||
| + | |||
| + | Target products exist in almost all samples. We selected No. 1, 2 colony for incubation for plasmid extraction and No. 6, 12, 14 for storing at 4C. | ||
| + | |||
| + | <br style="line-height: 0; clear: both;" /> | ||
</div></div> | </div></div> | ||
Latest revision as of 03:24, 27 September 2012
September 17th
Digestion of RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 and ptet-pSB1A2
To make a construct of ptet-RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 with XbaI and PstI, ptet-pSB1A2 with SpeI and PstI.
Insert (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2)
| DNA solution (45~50 ng/ul) | 27 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 4 ul |
| DW | 7 ul |
| Total | 40 ul |
Vector(ptet-pSB1A2)
| DNA solution (about 20 ng/ul) | 4 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 12 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 70 | 20 |
| 3 | 4 | HOLD |
Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We confirmed that the concentration of Insert DNA solution is 60~70 ng/ul and Vector DNA solution is about 20~30 ng/ul.
Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
| Insert DNA (60~70 ng/ul) | 4 ul |
| Vector DNA (20~30 ng/ul) | 2 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 12 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2
Transformation of ligation ligation product into JM109.
- Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBA).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 17 hrs.
September 18th
Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3
We did colony PCR two times.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(200bp down primer) | 0.8 ul |
| Reverse Primer(ag43-f4 primer) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.0 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls.
Desired product is about 702bp.
Target products didn't exist in all samples. We noticed the reason why such results shown is that we used incorrect pSB1C3 DNA solution which isn't confirmed the sequence. We'll try the synthesis once more.
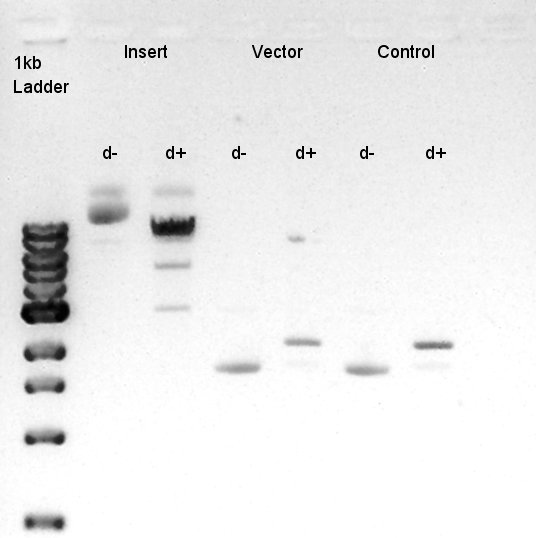
Digestion of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 and pSB1C3
To make a construct of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3, we digested pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 with EcoRI and SpeI and pSB1C3 with EcoRI and SpeI.
Insert (pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28)
| DNA solution (about 35ng/ul) | 41 ul |
| EcoRI | 2 ul |
| SpeI | 2 ul |
| 10xH buffer | 5 ul |
| Total | 50 ul |
Vector(pSB1C3)
| DNA solution (about 30~40 ng/ul) | 2 ul |
| EcoRI | 1 ul |
| SpeI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 14 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Ethanol precipitation of pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
Ligation of pBAD-RBS-eCFP-RBS-Ag43-dT and pSB1C3
| Insert DNA | 4 ul |
| Vector DNA | 2 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 12 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of pBAD-RBS-eCFP-RBS-Ag43-dT-pSB1C3
Transformation ligation product into JM109.
- Mixed 2 ul pBAD-RBS-eCFP-RBS-Ag43-dT--pSB1C3 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Incubated for 2 hrs to get the resistance to Chloramphenicol.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBC).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 15 hrs.
September 19th
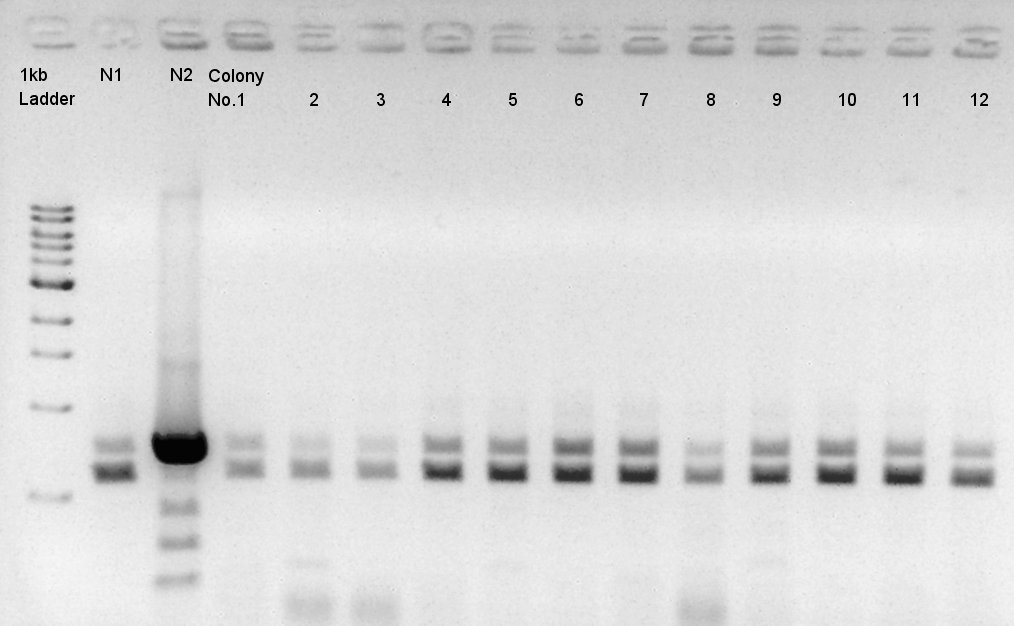
Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28
Colony PCR to confirm whether pSTV28 vettor has pBAS-RBs-eCFP-RBs-Ag43-dT as insert or not.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(200bp down primer) | 0.8 ul |
| Reverse Primer(ag43-f4 primer) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.3 | 30 |
| 4 | 72 | 180 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2(pBAD-RBS-Ag43-dT-pSB1A2)as controls.
Desired product is about 702bp.
We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA.
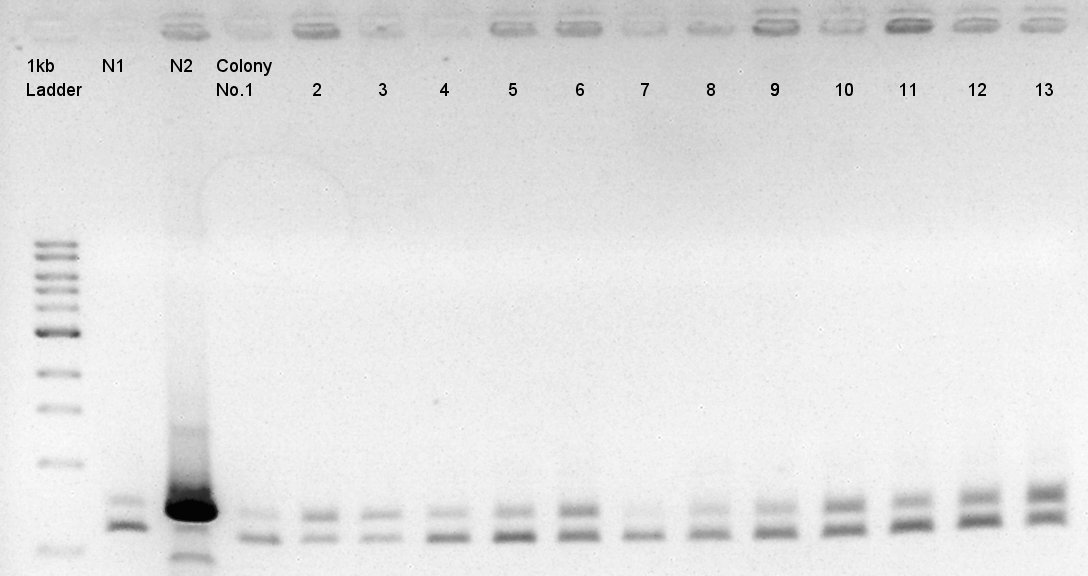
Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3 and pBAD-RBS-eCFP-RBS-Ag43-dI on pSB1C3
Colony PCR to confirm whether the constructs written above really have eCFP and Ag43 coding sites or not by using primers: one can anneal to a part of eCFP coding site and another can anneal to a part of Ag43 coding site.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(FP-F primer) | 0.8 ul |
| Reverse Primer(ag43-R primer) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.3 | 30 |
| 4 | 72 | 180 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1, N2 (DW only) as controls. Desired product is about 452 bp and 697 bp respectively.
We noticed the some of colones have pBAD-RBS-eCFP-RBS-Ag43-dT-pSTV28 construct as plasmid DNA.
September 20th
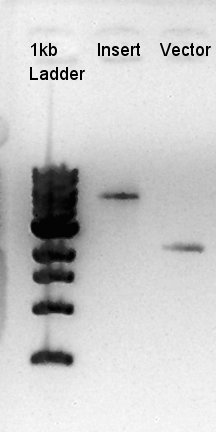
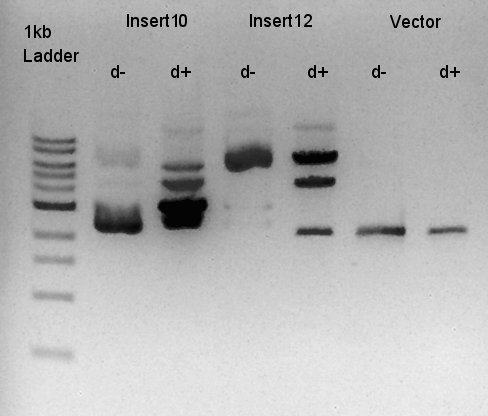
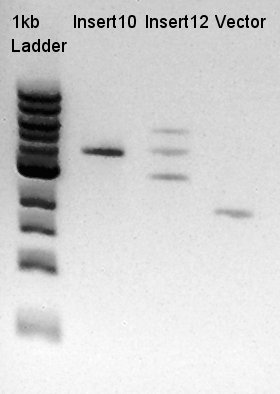
Digestion of ptet-RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 (DNA solution eluted from colony No.10, 12 respectively in ) and ptet-pSB1A2
To make a construct of ptet-RBS-phaC-RBS-phaA-RBS-phaB-RBS-dT-pSB1A2, we digested RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 with XbaI and PstI, ptet-pSB1A2 with SpeI and PstI. We prepared DNa solution derived from No. 10 and No. 12 colony selected selected by the result of colony PCR for 18th.
Insert No.10 (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2)
| DNA solution (40 ng/ul) | 20 ul |
| EcoRI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 4 ul |
| DW | 4 ul |
| Total | 30 ul |
Insert No.12 (RBS-phaC-RBs-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2)
| DNA solution (40 ng/ul) | 20 ul |
| EcoRI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 4 ul |
| DW | 4 ul |
| Total | 30 ul |
Vector(ptet-pSB1A2)
| DNA solution (about 40 ng/ul) | 2 ul |
| EcoRI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2 ul |
| DW | 14 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 120 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
Ethanol precipitation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged at 15000 rpm, 15 min at 4C.
- Removed supernatant and added 220 ul of 70% ethanol.
- Centrifuged at 15000 rpm, 10 min at 4C.
- Removed supernatant and dried out at room temperature, after that added 10 ul of DW.
We decided to use No. 10 digestion product for ligation, and confirmed that the concentration of Insert DNA solution is 50 ng/ul and Vector DNA solution is about 20~30 ng/ul.
Ligation of RBS-phaB-RBS-eYFP-dT and RBS-phaC-RBS-phaA-pSB1A2
| Insert DNA (50 ng/ul) | 4 ul |
| Vector DNA (20~30 ng/ul) | 1.5 ul |
| DW | 0.5 ul |
| Ligation Mighty Mix | 6 ul |
| Total | 12 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation of RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2
Transformation of ligation product into JM109.
- Mixed 2 ul RBS-phaC-RBS-phaA-RBS-phaB-RBS-eYFP-dT-pSB1A2 ligation product to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Mixed 350 ul of LB.
- Incubated for 2 hrs to get the resistance to Chloramphenicol.
- Prepared and Labeled two plastic plates with LB plate medium which contained appropriate antibiotics (LBC).
- Plated 300 ul of the culture onto first dish and spread.
- Mixed 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for 15 hrs.
September 21th
Colony PCR of pBAD-RBS-eCFP-RBS-Ag43-dI-pSB1C3
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 10 ul |
| Forward Primer(X-phaB-F primer) | 0.8 ul |
| Reverse Primer(PS-R primer) | 0.8 ul |
| DW | 4.4 ul |
| Total | 20 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 68.9 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2(RBS-phaC-RBs-phaA-RBS-phaB-pSB1A2)as controls.
Desired product is about 1500bp.
Target products exist in almost all samples. We selected No. 1, 2 colony for incubation for plasmid extraction and No. 6, 12, 14 for storing at 4C.
 "
"