Team:Bielefeld-Germany/Results/london
From 2012.igem.org
JSchirmacher (Talk | contribs) (→Laccase activity tests) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Bielefeld/Head}} | {{Team:Bielefeld/Head}} | ||
<html> | <html> | ||
| + | <a href="https://2012.igem.org/Team:Bielefeld-Germany/Results/Summary#8"><img src="http://2012.igem-bielefeld.de/includes/wiki/images/Pfeil_links2.png"></a> | ||
<div id=page-title> | <div id=page-title> | ||
<span id=page-title-text> | <span id=page-title-text> | ||
| - | Collaboration with <a href="https://2012.igem.org/Team:University_College_London"> | + | Collaboration with <a href="https://2012.igem.org/Team:University_College_London"> University College London</a> |
</span> | </span> | ||
</div> | </div> | ||
| Line 15: | Line 16: | ||
==Introduction== | ==Introduction== | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
| - | + | At the [https://2012.igem.org/Team:Bielefeld-Germany/Human_Practices/CAS_conference CAS conference] in Munich we met the iGEM team [https://2012.igem.org/Team:University_College_London UCL] for the first time. Discussing about the different projects a common basis was found. Since the UCL team also produced laccases (for a different approach) a collaboration has been developed subsequently. The idea was that each team will characterize the laccase(s) of the other team to reproduce and cross-validate the results. Therefore the plasmids containing specific BioBricks were exchanged. The iGEM UCL characterized our BioBrick <partinfo>K863005</partinfo> and consequently we characterized their BioBrick <partinfo>K729006</partinfo>. The laccase CueO from ''E. coli'' BL21(DE3) was used by both teams, but with different expression vectors and chassis as well as a different protocol for the laccase activity test . Hence, the established methods for production and cell disruption had to be discussed and adjusted. For the characterization of the activity each team used its own method. | |
| - | For | + | For an initial characterization the BioBrick <partinfo>K729006</partinfo> was sequenced. Afterwards, the protein was produced in ''E. coli'' KRX and a growth kinetic was determined. Furthermore, a SDS-PAGE as well as an activity test was performed. |
</div> | </div> | ||
==Sequencing analysis== | ==Sequencing analysis== | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
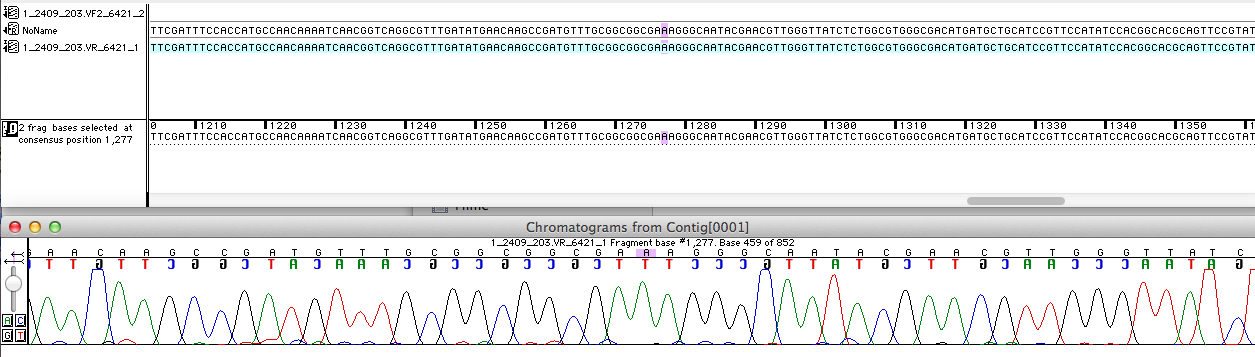
| - | For the improvement of the data collection | + | For the improvement of the data collection the BioBrick of the University College London iGEM team was sequenced. The sequence analysis shows that the analyzed BioBrick matches the sequence published in the Partsregistry (see Fig. 1). It was proven that the BioBrick codes for the ''E. coli'' laccase CueO, indeed. Therefore the sequence was aligned with CueO from ''E. coli'' BL21(DE3). It matched with 100 %. |
| - | [[File:Bielefeld2012_Sequencher.png|700px|thumb|center|'''Figure 1:''' A short part of the sequencing analysis results of the BioBrick <partinfo>K729006</partinfo> ]] | + | [[File:Bielefeld2012_Sequencher.png|700px|thumb|center|'''Figure 1:''' A short part of the sequencing analysis results of the BioBrick <partinfo>K729006</partinfo>. ]] |
</div> | </div> | ||
==Cultivation== | ==Cultivation== | ||
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
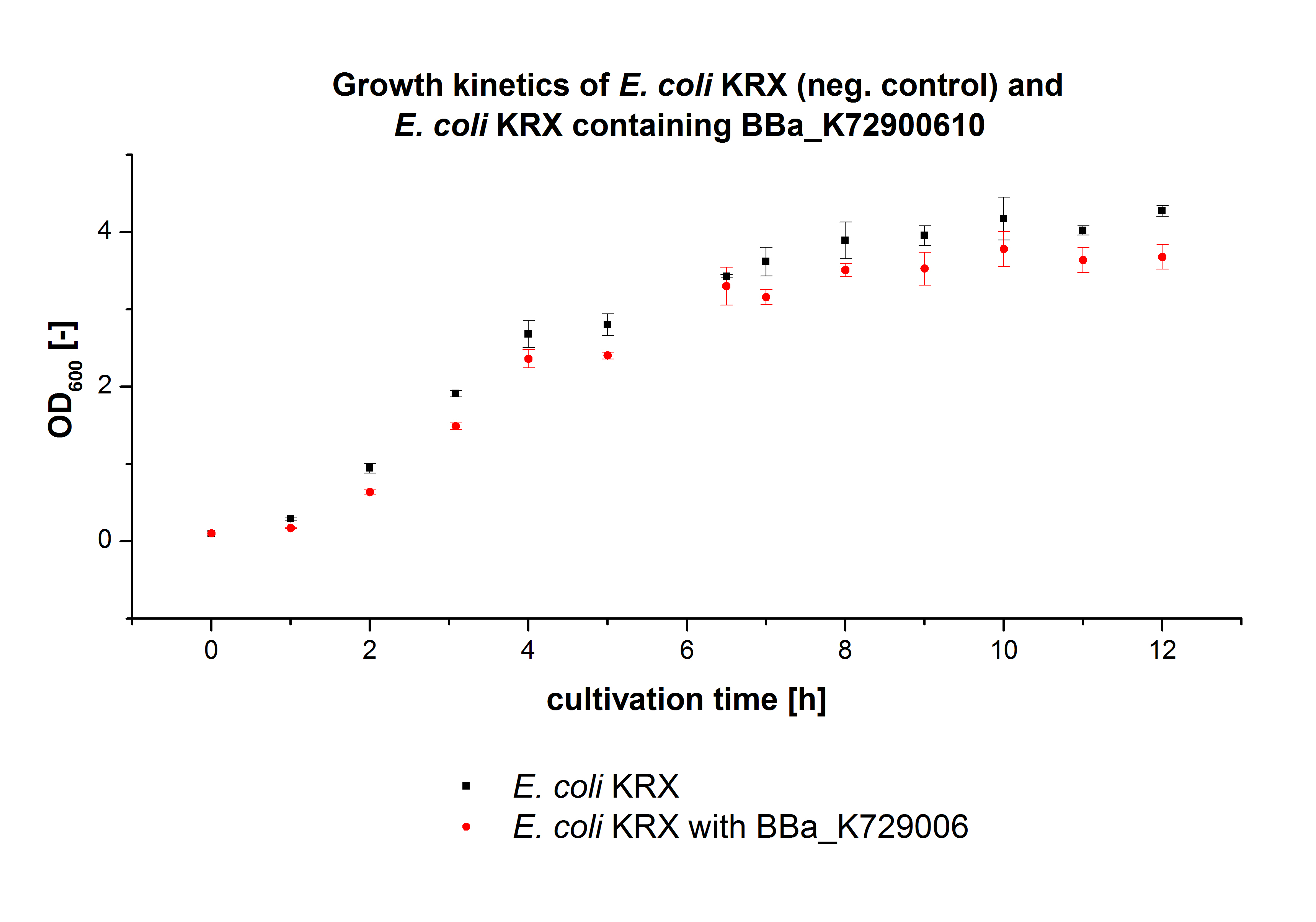
| - | [[File:Bielefeld2012_Wachstumskinetik_pLondon.jpg|450px|thumb|left| '''Figure 2:''' Comparison of the cultivated ''E. coli'' KRX and ''E. coli'' KRX containing <partinfo>K729006</partinfo> cultivations. The OD<html><sub>600</sub></html> was measured | + | [[File:Bielefeld2012_Wachstumskinetik_pLondon.jpg|450px|thumb|left| '''Figure 2:''' Comparison of the cultivated ''E. coli'' KRX and ''E. coli'' KRX containing <partinfo>K729006</partinfo> cultivations. The OD<html><sub>600</sub></html> was measured each hour for 12 h.]] |
| - | ''E. coli'' KRX was transformed with the BioBrick <partinfo>K729006</partinfo> of the | + | ''E. coli'' KRX was transformed with the BioBrick <partinfo>K729006</partinfo> of the UCL team. The cells were characterized by growth kinetics and activity test. |
| - | ''E. coli'' KRX containing <partinfo>K729006</partinfo> was cultivated in [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB-medium] with a total volume of 60 mL for 12 h at 37 °C (120 rpm) in 250 mL shaking flasks without baffles. To produce a higher amount of the protein | + | ''E. coli'' KRX containing <partinfo>K729006</partinfo> was cultivated in [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB-medium] with a total volume of 60 mL for 12 h at 37 °C (120 rpm) in 250 mL shaking flasks without baffles. To produce a higher amount of the protein we also cultivated a total volume of 200 mL in a 1 L shaking flask without baffles. The OD<html><sub>600</sub></html> values were determined each hour. To measure the influence of the transformed BioBrick on the growth of the cells a negative control (''E. coli'' KRX) was cultivated identically. |
| - | In | + | In Fig. 2 the growth curves of''E. coli'' KRX containing <partinfo>K729006</partinfo> and of the negative control ''E. coli'' KRX are plotted. As expected the ''E. coli'' KRX containing <partinfo>K729006</partinfo> grew slower than the negative control. This can be explained by the protein expression. |
| - | The cells were harvested and centrifuged after 12 h. The pellets were resuspended in 100 mM Na-Acetat-buffer, lysed by [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis | + | The cells were harvested and centrifuged after 12 h. The pellets were resuspended in 100 mM Na-Acetat-buffer, lysed by [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Production#Mechanical_lysis sonication] and centrifuged. The supernatant was analyzed by SDS-PAGE and laccase activity assay. |
</div> | </div> | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 43: | Line 44: | ||
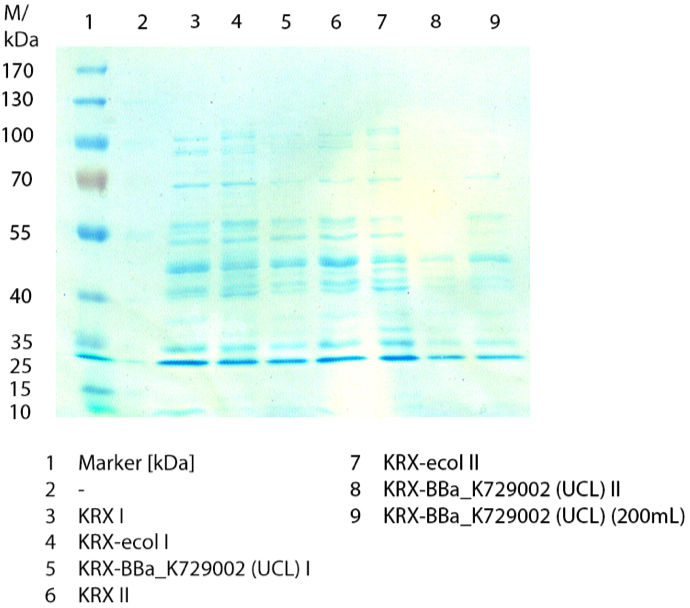
[[File:Bielefeld2012_0924.jpg|300px|thumb|left|'''Figure 3:''' SDS-PAGE of the lysates of ''E. coli'' KRX with <partinfo>BBa_K729006</partinfo>, ''E. coli'' KRX (negative control) and with <partinfo>BBa_K863005</partinfo> (positive control). ECOL has a molecular weight of 53.4 kDa. ]] | [[File:Bielefeld2012_0924.jpg|300px|thumb|left|'''Figure 3:''' SDS-PAGE of the lysates of ''E. coli'' KRX with <partinfo>BBa_K729006</partinfo>, ''E. coli'' KRX (negative control) and with <partinfo>BBa_K863005</partinfo> (positive control). ECOL has a molecular weight of 53.4 kDa. ]] | ||
| - | In | + | In Fig. 3 the SDS-PAGE of the ''E. coli'' KRX lysates with <partinfo>BBa_K729006</partinfo>, <partinfo>BBa_K863005</partinfo> and without any BioBrick (negative control) are shown after they have been treated the same way. No clear distinction between the samples is possible. By comparing the bands at 53.4 kDa (which matches the molecular weight of ECOL) just a small difference between the intensities of the bands can be considered. The ''E. coli'' KRX containing <partinfo>BBa_K863005</partinfo> has a slightly stronger band. |
</div> | </div> | ||
<br style="clear: both" /> | <br style="clear: both" /> | ||
| - | == | + | ==Laccase activity tests== |
<div style="text-align:justify;"> | <div style="text-align:justify;"> | ||
| - | + | Laccase activity tests were performed by using 198 µL of the sample (supernatant of the lysed cells containing <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo>) and 2 µL ABTS (20 mM ABTS stock). Since laccases are capable of oxidizing ABTS the activity of laccases can be detected by measuring the absorption of oxidized ABTS at 420 nm. As a positive control the ''E. coli'' KRX strain containing <partinfo>K863005</partinfo> which already has been [https://2012.igem.org/Team:Bielefeld-Germany/Results/Summary#4 characterized] by the team was applied as well. Using this approach there was no detectable activity over a period of 40 minutes (see Fig. 4). To gain activity the samples were incubated with 0.4 mM CuCl<html><sub>2</sub></html> for at least 2 hours. After incubation time another setting was started using 198 µL of the samples containing <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> and applied 0.1 mM ABTS. The measurements lasted for 5 hours but the results did not show a decrease in oxidized ABTS again (see Fig. 5). A possible reason can be contaminants in the samples, e.g. salts or low molecular agents, that could have affected the activity of the tested laccases. The samples were rebuffered in deionized H<sub>2</sub>O using HiTrap Desalting Columns and then again incubated with 0.4 mM CuCl<html><sub>2</sub></html> for 2 hours. This time the measurement setup differed because no buffer remained in the samples. The new measurements were performed with 100 mM sodium acetate buffer (pH 5), 158 µL sample and 0.01 mM ABTS for 5 hours at a temperature of 25°C. This sort of sample preparation led to oxidizing activity of the laccases and therefore to an increase in oxidized ABTS (see Fig. 6). After 5 hours both laccases <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> showed high activity and oxidized around 8 µM ABTS. Since both laccases derive from the same organism they both show a similar reaction behavior. | |
</div> | </div> | ||
<center> | <center> | ||
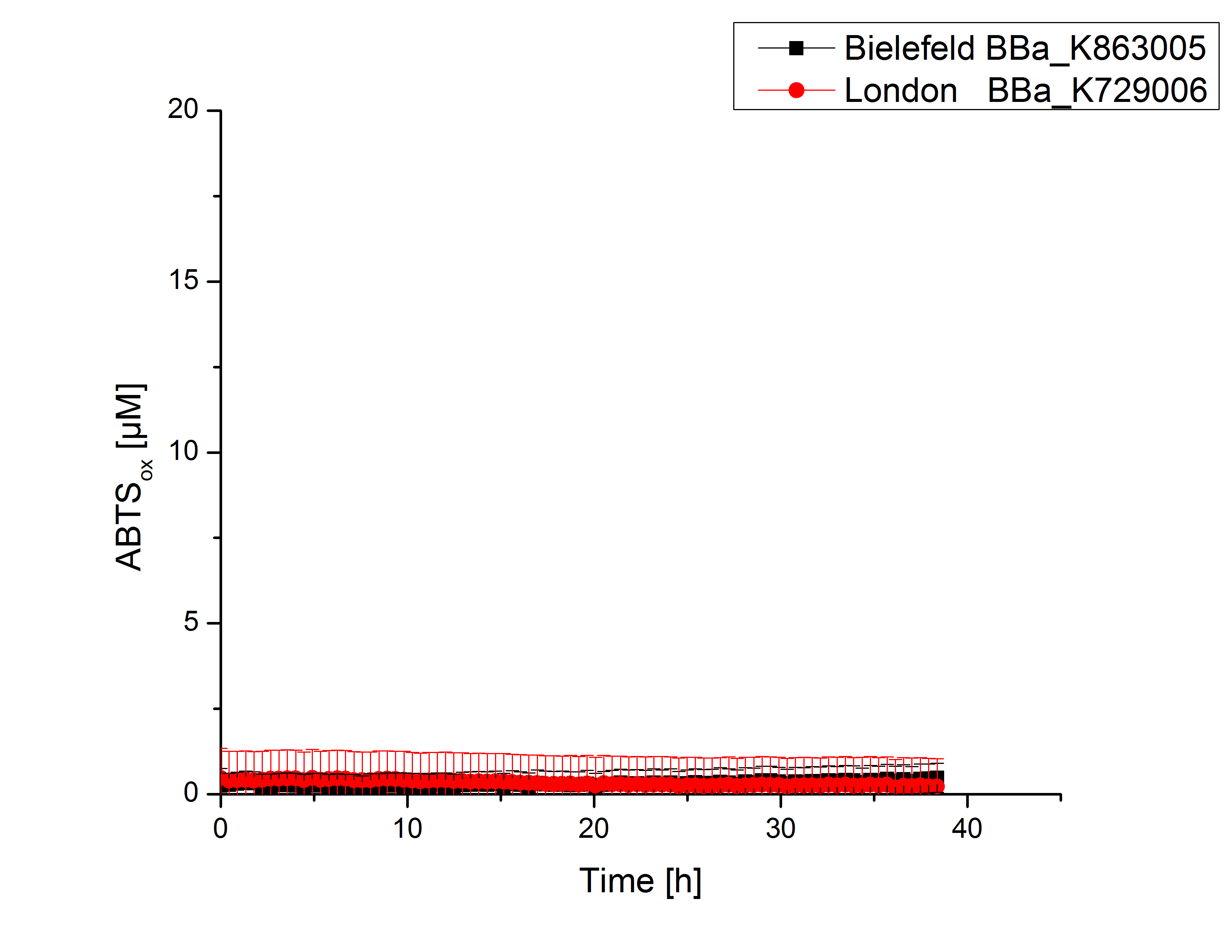
| - | [[File:Bielefeld2012_London_ohnecu_mitsalz.jpg|240px|left|thumbnail|'''Figure 4:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> using ABTS over a time period of 40 minutes. No increase in oxidized ABTS | + | [[File:Bielefeld2012_London_ohnecu_mitsalz.jpg|240px|left|thumbnail|'''Figure 4:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> using ABTS over a time period of 40 minutes. No increase in oxidized ABTS was detectable.]] |
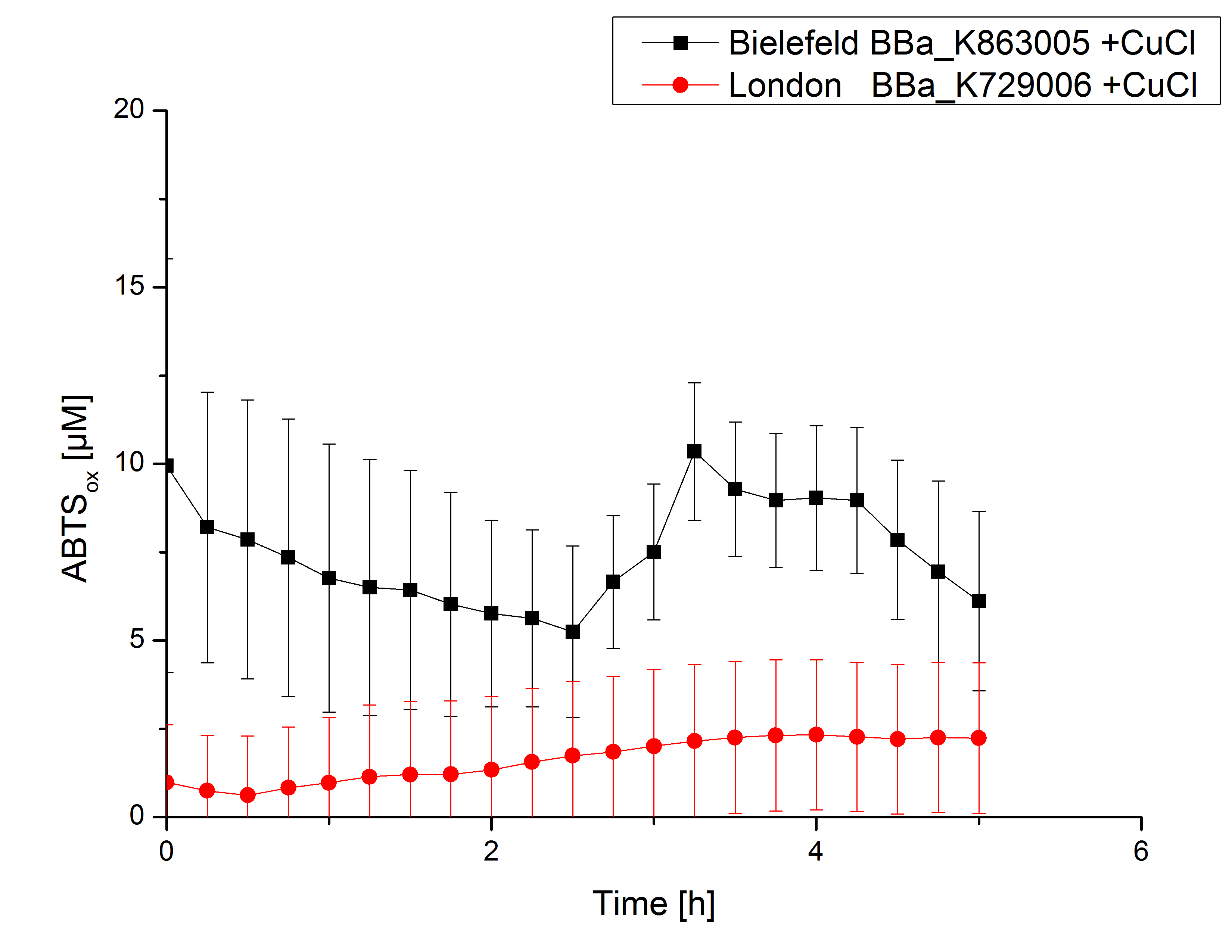
| - | [[File:Bielefeld2012_London_cu.jpg|240px|left|thumbnail|'''Figure 5:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> after | + | [[File:Bielefeld2012_London_cu.jpg|240px|left|thumbnail|'''Figure 5:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> after 2 hours incubation with 0.4 mM CuCl<sub>2</sub> using ABTS over a time period of 5 hours. No activity of both laccases could be detected since there was no increase in oxidized ABTS.]] |
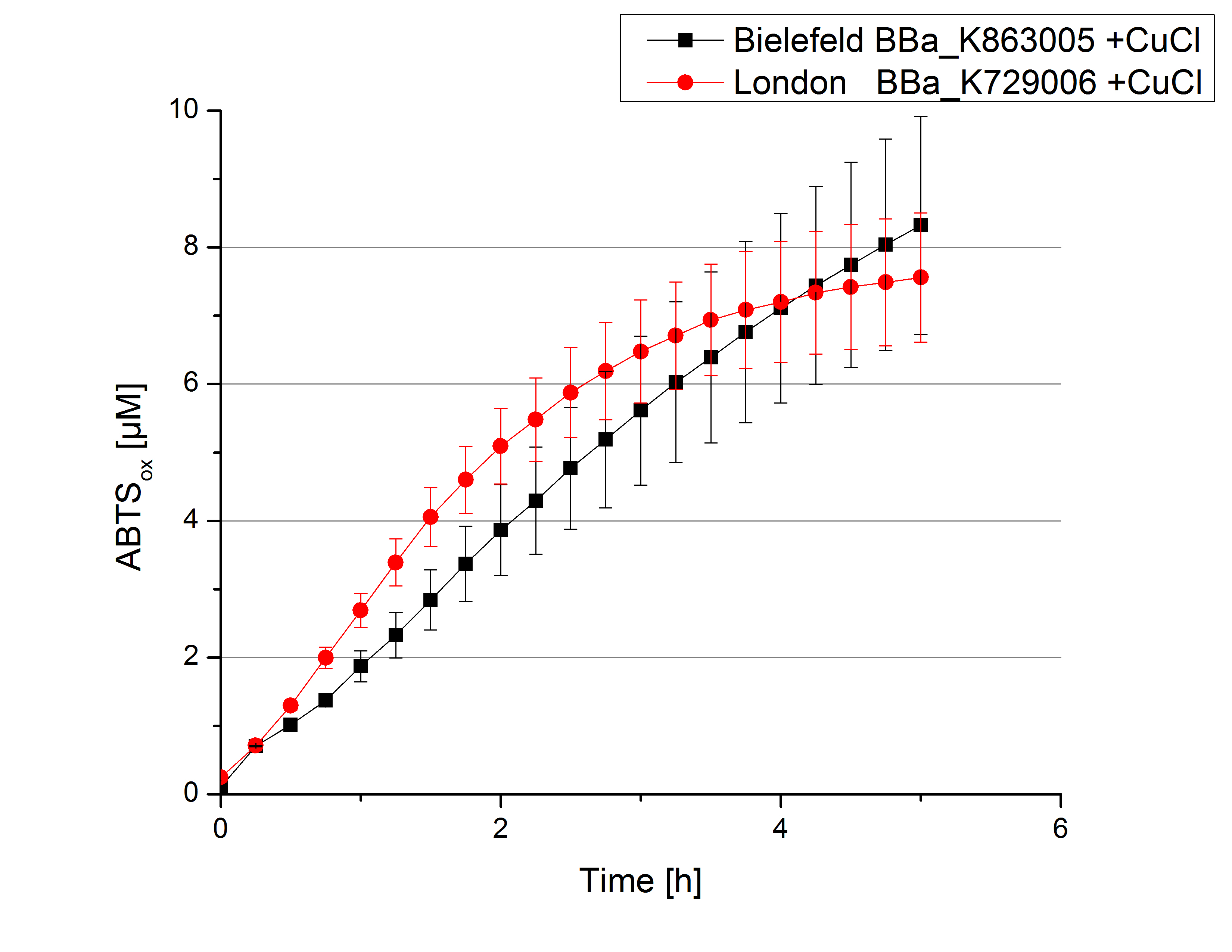
| - | [[File:Bielefeld2012_London_umgepuffert_cu.jpg|240px|left|thumbnail|'''Figure 6:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> after desalting and | + | [[File:Bielefeld2012_London_umgepuffert_cu.jpg|240px|left|thumbnail|'''Figure 6:''' Activity measurement of <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> after desalting and 2 hours incubation with 0.4 mM CuCl<sub>2</sub> using ABTS over a time period of 5 hours. A considerable increase in oxidized ABTS is noticeable. After 5 hours both laccases were capable of oxidizing 8 µM ABTS.]] |
</center> | </center> | ||
Latest revision as of 00:55, 27 September 2012

Summary
Contents |
Introduction
At the CAS conference in Munich we met the iGEM team UCL for the first time. Discussing about the different projects a common basis was found. Since the UCL team also produced laccases (for a different approach) a collaboration has been developed subsequently. The idea was that each team will characterize the laccase(s) of the other team to reproduce and cross-validate the results. Therefore the plasmids containing specific BioBricks were exchanged. The iGEM UCL characterized our BioBrick <partinfo>K863005</partinfo> and consequently we characterized their BioBrick <partinfo>K729006</partinfo>. The laccase CueO from E. coli BL21(DE3) was used by both teams, but with different expression vectors and chassis as well as a different protocol for the laccase activity test . Hence, the established methods for production and cell disruption had to be discussed and adjusted. For the characterization of the activity each team used its own method.
For an initial characterization the BioBrick <partinfo>K729006</partinfo> was sequenced. Afterwards, the protein was produced in E. coli KRX and a growth kinetic was determined. Furthermore, a SDS-PAGE as well as an activity test was performed.
Sequencing analysis
For the improvement of the data collection the BioBrick of the University College London iGEM team was sequenced. The sequence analysis shows that the analyzed BioBrick matches the sequence published in the Partsregistry (see Fig. 1). It was proven that the BioBrick codes for the E. coli laccase CueO, indeed. Therefore the sequence was aligned with CueO from E. coli BL21(DE3). It matched with 100 %.
Cultivation
E. coli KRX was transformed with the BioBrick <partinfo>K729006</partinfo> of the UCL team. The cells were characterized by growth kinetics and activity test. E. coli KRX containing <partinfo>K729006</partinfo> was cultivated in LB-medium with a total volume of 60 mL for 12 h at 37 °C (120 rpm) in 250 mL shaking flasks without baffles. To produce a higher amount of the protein we also cultivated a total volume of 200 mL in a 1 L shaking flask without baffles. The OD600 values were determined each hour. To measure the influence of the transformed BioBrick on the growth of the cells a negative control (E. coli KRX) was cultivated identically.
In Fig. 2 the growth curves ofE. coli KRX containing <partinfo>K729006</partinfo> and of the negative control E. coli KRX are plotted. As expected the E. coli KRX containing <partinfo>K729006</partinfo> grew slower than the negative control. This can be explained by the protein expression.
The cells were harvested and centrifuged after 12 h. The pellets were resuspended in 100 mM Na-Acetat-buffer, lysed by sonication and centrifuged. The supernatant was analyzed by SDS-PAGE and laccase activity assay.
SDS-PAGE
In Fig. 3 the SDS-PAGE of the E. coli KRX lysates with <partinfo>BBa_K729006</partinfo>, <partinfo>BBa_K863005</partinfo> and without any BioBrick (negative control) are shown after they have been treated the same way. No clear distinction between the samples is possible. By comparing the bands at 53.4 kDa (which matches the molecular weight of ECOL) just a small difference between the intensities of the bands can be considered. The E. coli KRX containing <partinfo>BBa_K863005</partinfo> has a slightly stronger band.
Laccase activity tests
Laccase activity tests were performed by using 198 µL of the sample (supernatant of the lysed cells containing <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo>) and 2 µL ABTS (20 mM ABTS stock). Since laccases are capable of oxidizing ABTS the activity of laccases can be detected by measuring the absorption of oxidized ABTS at 420 nm. As a positive control the E. coli KRX strain containing <partinfo>K863005</partinfo> which already has been characterized by the team was applied as well. Using this approach there was no detectable activity over a period of 40 minutes (see Fig. 4). To gain activity the samples were incubated with 0.4 mM CuCl2 for at least 2 hours. After incubation time another setting was started using 198 µL of the samples containing <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> and applied 0.1 mM ABTS. The measurements lasted for 5 hours but the results did not show a decrease in oxidized ABTS again (see Fig. 5). A possible reason can be contaminants in the samples, e.g. salts or low molecular agents, that could have affected the activity of the tested laccases. The samples were rebuffered in deionized H2O using HiTrap Desalting Columns and then again incubated with 0.4 mM CuCl2 for 2 hours. This time the measurement setup differed because no buffer remained in the samples. The new measurements were performed with 100 mM sodium acetate buffer (pH 5), 158 µL sample and 0.01 mM ABTS for 5 hours at a temperature of 25°C. This sort of sample preparation led to oxidizing activity of the laccases and therefore to an increase in oxidized ABTS (see Fig. 6). After 5 hours both laccases <partinfo>K729006</partinfo> and <partinfo>K863005</partinfo> showed high activity and oxidized around 8 µM ABTS. Since both laccases derive from the same organism they both show a similar reaction behavior.
| 55px | | | | | | | | | | |
 "
"