Team:Paris Bettencourt/Restriction Enzyme
From 2012.igem.org
(→Design) |
(→Design) |
||
| Line 59: | Line 59: | ||
<ul> | <ul> | ||
<li> | <li> | ||
| - | Standard pLac promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_R0011 R0011].<sup>[</sup><sup>[[#References| | + | Standard pLac promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_R0011 R0011].<sup>[</sup><sup>[[#References|7]]</sup><sup>]</sup> |
</li> | </li> | ||
</ul> | </ul> | ||
| Line 67: | Line 67: | ||
<br/> | <br/> | ||
<ul> | <ul> | ||
| - | <li>Standard pBad promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_I13453 I13453].</li> | + | <li>Standard pBad promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_I13453 I13453].<sup>[</sup><sup>[[#References|8]]</sup><sup>]</sup></li> |
</ul> | </ul> | ||
</li> | </li> | ||
| Line 84: | Line 84: | ||
</li> | </li> | ||
| - | <li>L-rhamnose is taken up by the RhaT transport system, converted to L-rhamnulose by an isomerase RhaA and then phosphorylated by a kinase RhaB. Subsequently, the resulting rhamnulose-1-phosphate is hydrolyzed by an aldolase RhaD into dihydroxyacetone phosphate, which is metabolized in glycolysis, and L-lactaldehyde. The latter can be oxidized into lactate under aerobic conditions and be reduced into L-1,2-propanediol under unaerobic conditions.</li> | + | <li>L-rhamnose is taken up by the RhaT transport system, converted to L-rhamnulose by an isomerase RhaA and then phosphorylated by a kinase RhaB. Subsequently, the resulting rhamnulose-1-phosphate is hydrolyzed by an aldolase RhaD into dihydroxyacetone phosphate, which is metabolized in glycolysis, and L-lactaldehyde. The latter can be oxidized into lactate under aerobic conditions and be reduced into L-1,2-propanediol under unaerobic conditions.<sup>[</sup><sup>[[#References|7]]</sup><sup>]</sup></li> |
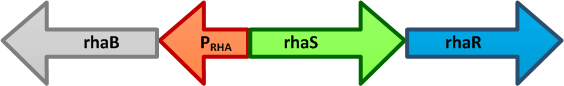
| - | [[Image:Paris_Bettencourt_2012_Prha.png|thumb|400px|right|alt=The E. coli rhaBRS locus|<font size="1"><b>The ''E. coli'' rhaBRS locus.</b> In the presence of | + | [[Image:Paris_Bettencourt_2012_Prha.png|thumb|400px|right|alt=The E. coli rhaBRS locus|<font size="1"><b>The ''E. coli'' rhaBRS locus.</b> In the presence of L-rhamnose, RhaR activates transcription of ''rhaR'' and ''rhaS'', resulting in an accumulation of RhaS. RhaS then acts as the L-rhamnose-dependent positive regulator of the ''rhaB'' promoter.</font>]] |
| - | <li>The genes rhaBAD are organized in one operon which is controlled by the rhaPBAD promoter. This promoter is regulated by two activators, RhaS and RhaR, and the corresponding genes belong to one transcription unit which is located in opposite direction of rhaBAD. If L-rhamnose is available, RhaR binds to the rhaPRS promoter and activates the production of RhaR and RhaS. RhaS together with L-rhamnose in turn binds to the rhaPBAD and the rhaPT promoter and activates the transcription of the structural genes. However, for the application of the rhamnose expression system it is not necessary to express the regulatory proteins in larger quantities, because the amounts expressed from the chromosome are sufficient to activate transcription even on multi-copy plasmids. Therefore, only the rhaPBAD promoter has to be cloned upstream of the gene that is to be expressed. Full induction of rhaBAD transcription also requires binding of the CRP-cAMP complex, which is a key regulator of catabolite repression.</li> | + | <li>The genes rhaBAD are organized in one operon which is controlled by the rhaPBAD promoter. This promoter is regulated by two activators, RhaS and RhaR, and the corresponding genes belong to one transcription unit which is located in opposite direction of rhaBAD. If L-rhamnose is available, RhaR binds to the rhaPRS promoter and activates the production of RhaR and RhaS. RhaS together with L-rhamnose in turn binds to the rhaPBAD and the rhaPT promoter and activates the transcription of the structural genes. However, for the application of the rhamnose expression system it is not necessary to express the regulatory proteins in larger quantities, because the amounts expressed from the chromosome are sufficient to activate transcription even on multi-copy plasmids. Therefore, only the rhaPBAD promoter has to be cloned upstream of the gene that is to be expressed. Full induction of rhaBAD transcription also requires binding of the CRP-cAMP complex, which is a key regulator of catabolite repression.<sup>[</sup><sup>[[#References|11]]</sup><sup>]</sup></li> |
| - | <li>The pRha sequence was containing an EcoRI restriction site, so we had to disrupt it in order to use pRha as a biobrick. In order to decide which base pair to modify, we used the [http://microbes.ucsc.edu UCSC Microbial Genome Browser]. We compared the pRha sequence in E.coli and similar species, and identified that the at the position is sometimes replaced by a in some species, so we decided to replace it in a same way. We ordered a gBlock with the pRha sequence having the mutation, and this is the sequence we used and submitted. | + | <li>The pRha sequence was containing an EcoRI restriction site, so we had to disrupt it in order to use pRha as a biobrick. In order to decide which base pair to modify, we used the [http://microbes.ucsc.edu UCSC Microbial Genome Browser]. We compared the pRha sequence in E.coli and similar species, and identified that the at the position is sometimes replaced by a in some species, so we decided to replace it in a same way. We ordered a gBlock with the pRha sequence having the mutation, and this is the sequence we used and submitted.</li> |
</ul> | </ul> | ||
Revision as of 22:28, 26 September 2012
|
Achievements :
|
Contents |
Overview
Our group was responsible for designing a plasmid self-digestion system. This synthetic system allows to digest plasmids into linear parts of DNA and thus disrupt the expression of the genes carried by this plasmid, including the antitoxin (Colicin immunity protein), and any plugged-in synthetic device. Afterwards the cell's DNA can be degraded by the Colicin.
Objectives
- Find appropriate restriction enzymes which have to match the following properties:
- The corresponding restriction site must not be found in the E.Coli genome;
- The enzyme has to have high specifity;
- It has to work in wide range of different conditions (pH, T°, etc)
- Choose a strong yet tightly repressible promoter to regulate the restriction enzyme expression;
- Clone circuits with different combinations of the chosen restriction enzymes and promoters;
- Measure the degradation efficiency of the restriction enzyme for each circuit;
- Based on the best combination, design a self-disruption plasmid.
Design
According to the first two of our objectives, we should find an appropriate restriction enzyme and to choose a strong yet tightly repressible promoter to regulate its expression.
Restriction enzyme candidates:
- Fse I is a restriction endonuclease which recognizes an 8bp long DNA sequence: GGCCGG▽CC (CC△GGCCGG). The reason why we chose it is because it has the lowest number of restriction sites in the E.coli genome: only 4 copies. We decided to use MAGE to remove those sites from the chromosome (see for more details). However, MAGE did not have the expected yield, and we decided to freeze the work on this restriction enzyme and focus on the second candidate.
- I-SceI is an intron-encoded endonuclease. It is present in the mitochondria of Saccharomyces cerevisiae and recognises an 18-base pair sequence 5'-TAGGGATAA▽CAGGGTAAT-3' (3'-ATCCC△TATTGTCCCATTA-5') and leaves a 4 base pair 3' hydroxyl overhang. It is a rare cutting endonuclease. Statistically an 18-bp sequence will occur once in every 6.9*1010 base pairs or once in 20 human genomes.
Promoter candidates:
- pLac
- Standard pLac promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_R0011 R0011].[7]
- pBad
- Standard pBad promoter from the Parts Registry: [http://partsregistry.org/Part:BBa_I13453 I13453].[8]
- pRha L-rhamnose-inducible promoter is capable of high-level protein expression in the presence of L-rhamnose, it is also tightly regulated in the absence of L-rhamnose and by the addition of D-glucose.
- pRha is probably the best candidate for us because of two reasons:
- It is a new promoter, and so it will be orthogonal to any other existing system. It supports the modularity idea of our project.
- It was reported in the literature to be appropriate for the expression of toxic genes.
- L-rhamnose is taken up by the RhaT transport system, converted to L-rhamnulose by an isomerase RhaA and then phosphorylated by a kinase RhaB. Subsequently, the resulting rhamnulose-1-phosphate is hydrolyzed by an aldolase RhaD into dihydroxyacetone phosphate, which is metabolized in glycolysis, and L-lactaldehyde. The latter can be oxidized into lactate under aerobic conditions and be reduced into L-1,2-propanediol under unaerobic conditions.[7]
- The genes rhaBAD are organized in one operon which is controlled by the rhaPBAD promoter. This promoter is regulated by two activators, RhaS and RhaR, and the corresponding genes belong to one transcription unit which is located in opposite direction of rhaBAD. If L-rhamnose is available, RhaR binds to the rhaPRS promoter and activates the production of RhaR and RhaS. RhaS together with L-rhamnose in turn binds to the rhaPBAD and the rhaPT promoter and activates the transcription of the structural genes. However, for the application of the rhamnose expression system it is not necessary to express the regulatory proteins in larger quantities, because the amounts expressed from the chromosome are sufficient to activate transcription even on multi-copy plasmids. Therefore, only the rhaPBAD promoter has to be cloned upstream of the gene that is to be expressed. Full induction of rhaBAD transcription also requires binding of the CRP-cAMP complex, which is a key regulator of catabolite repression.[11]
- The pRha sequence was containing an EcoRI restriction site, so we had to disrupt it in order to use pRha as a biobrick. In order to decide which base pair to modify, we used the [http://microbes.ucsc.edu UCSC Microbial Genome Browser]. We compared the pRha sequence in E.coli and similar species, and identified that the at the position is sometimes replaced by a in some species, so we decided to replace it in a same way. We ordered a gBlock with the pRha sequence having the mutation, and this is the sequence we used and submitted.
- pRha is probably the best candidate for us because of two reasons:
Considering these candidates, we decided to clone the following constructs in low-copy vector pSB3C5 to use it in our experiments, all with a medium RBS [http://partsregistry.org/Part:BBa_B0032 B0032]:
| pBad & RBS & I-SceI | pRha & RBS & GFP | pBad & RBS & RFP |
| pLac & RBS & I-SceI | pRha & RBS & GFP | pLac & RBS & RFP |
| pRha & RBS & I-SceI | pRha & RBS & GFP | pRha & RBS & RFP |
Experiments and results
Measuring the efficiency of I-SceI (Cloned parts)
To measure the digestion efficiency of I-SceI, we did a trasformation of two plasmids with different antibiotic resistances into NEB Turbo E.Coli strain:
- First plasmid: Low copy plasmid with encoded generator to express I-SceI meganucllease. Three version with different promoters was tested: I-SceI meganuclease controlled by pBad, pLac and pRha. For all version:
- Backbone: pSB3C5
- Resistance: Chloramphenicol
- Origin of Replication: p15a
- Second plasmid: High copy plasmid with encoded I-SceI restriction site, [http://partsregistry.org/Part:BBa_K175027 K175027]. This biobrick was sent us by TUDelft iGEM team.
- Backbone: pSB1AK3
- Resistance: Ampicillin and Kanamycin
- Origin of Replication: modified pMB1 derived from pUC19
We expected to perform transformation with both plasmids, and plate with two antibiotics in order to select for double transformants. We would then induce I-SceI expression in those clones to measure its efficiency.
Transformation results
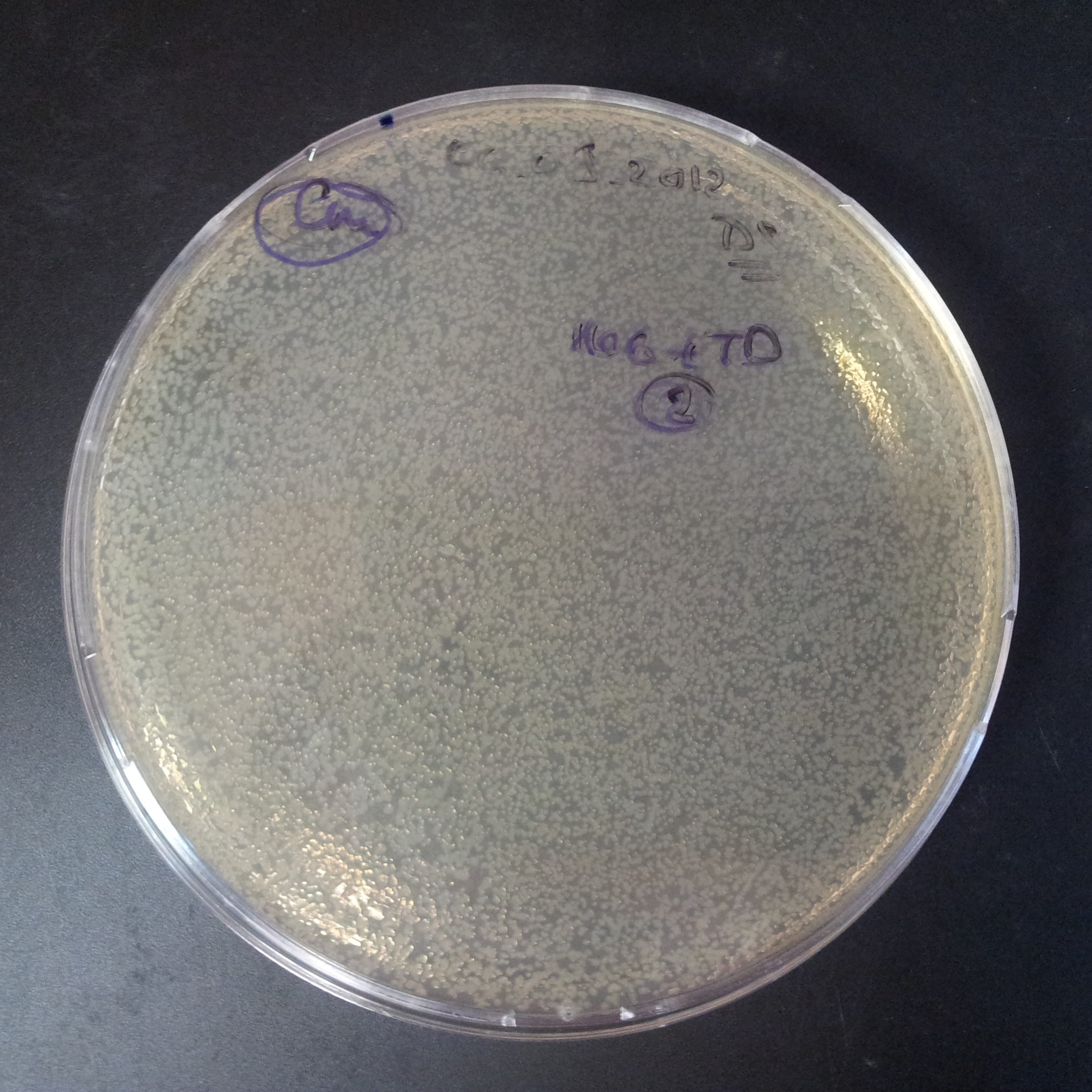
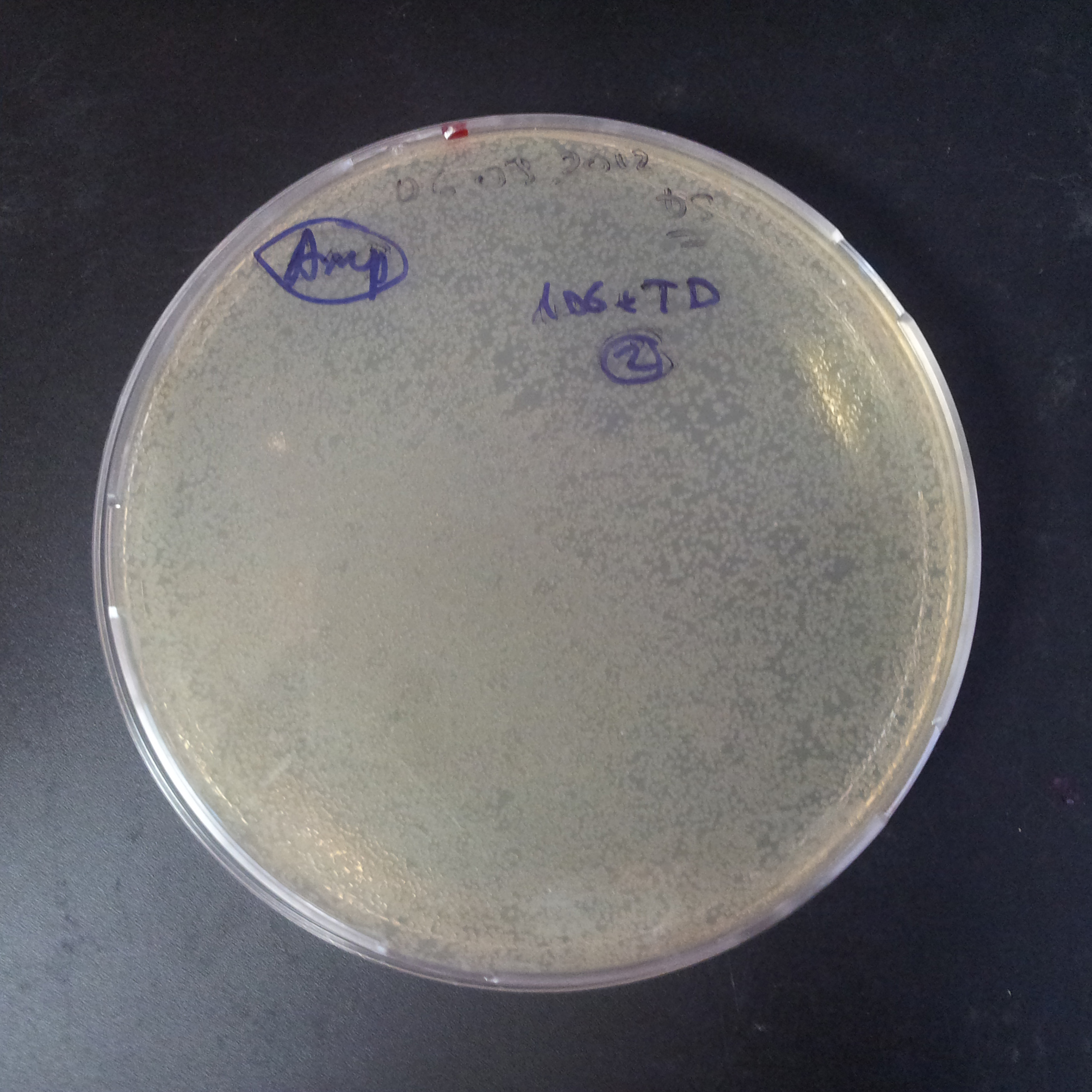
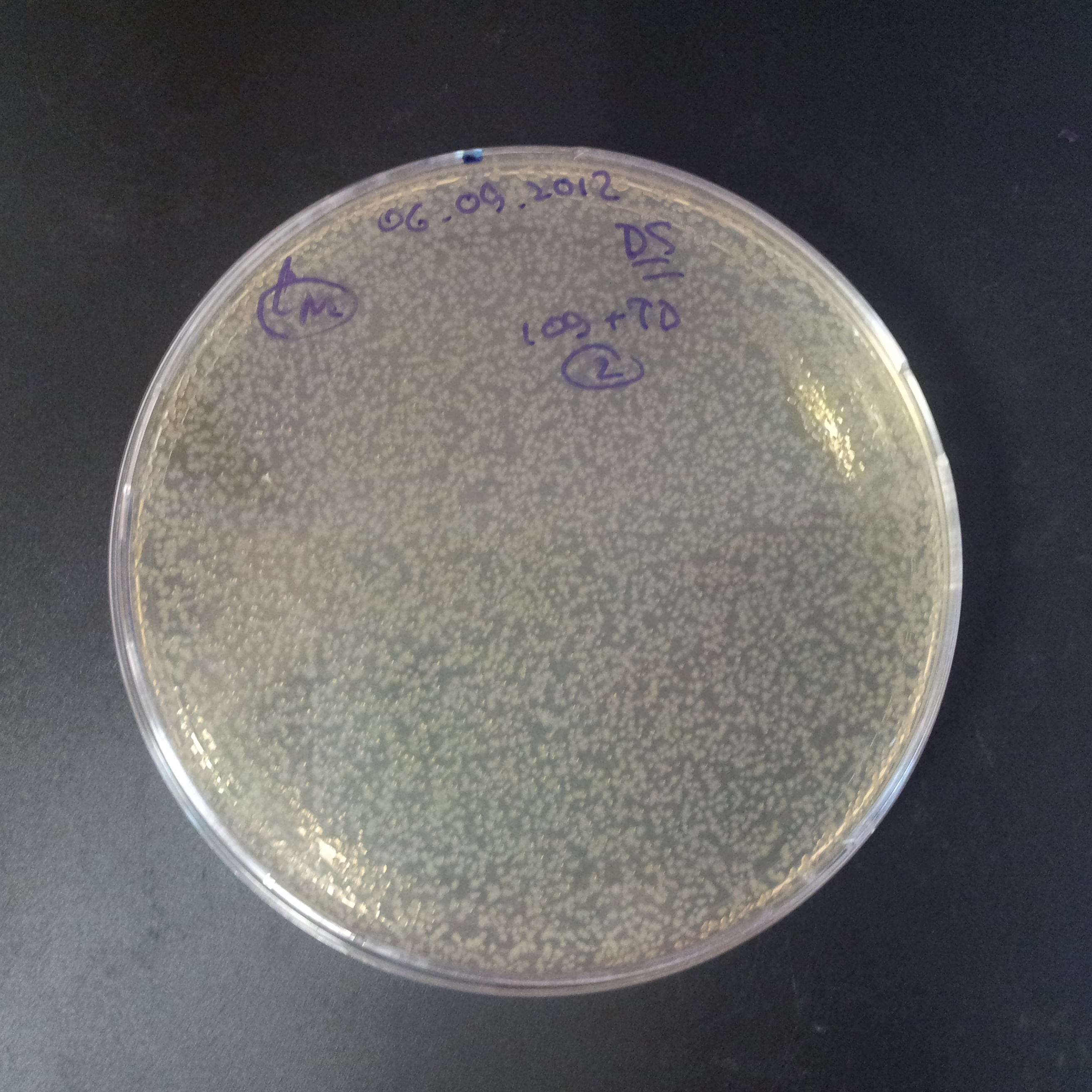
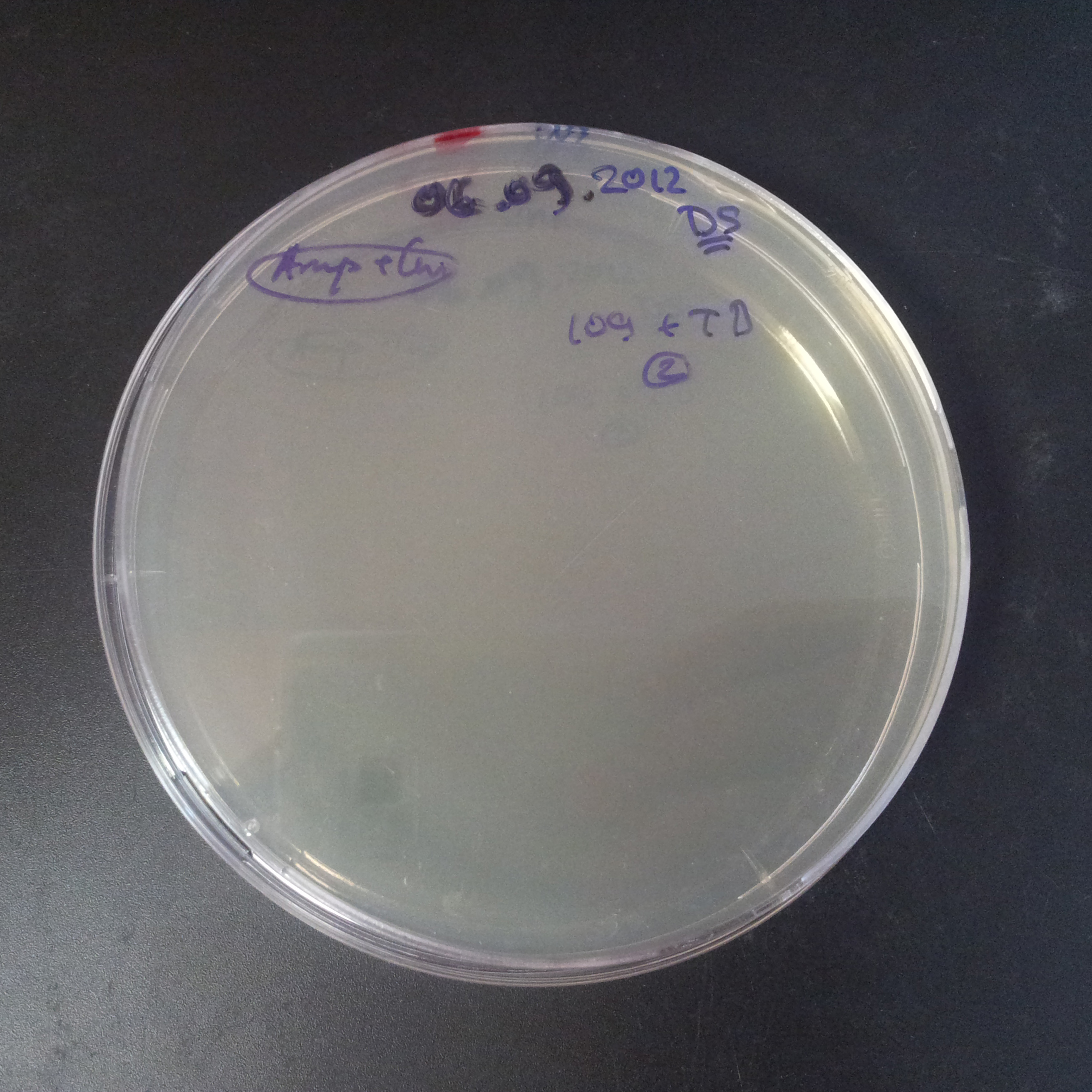
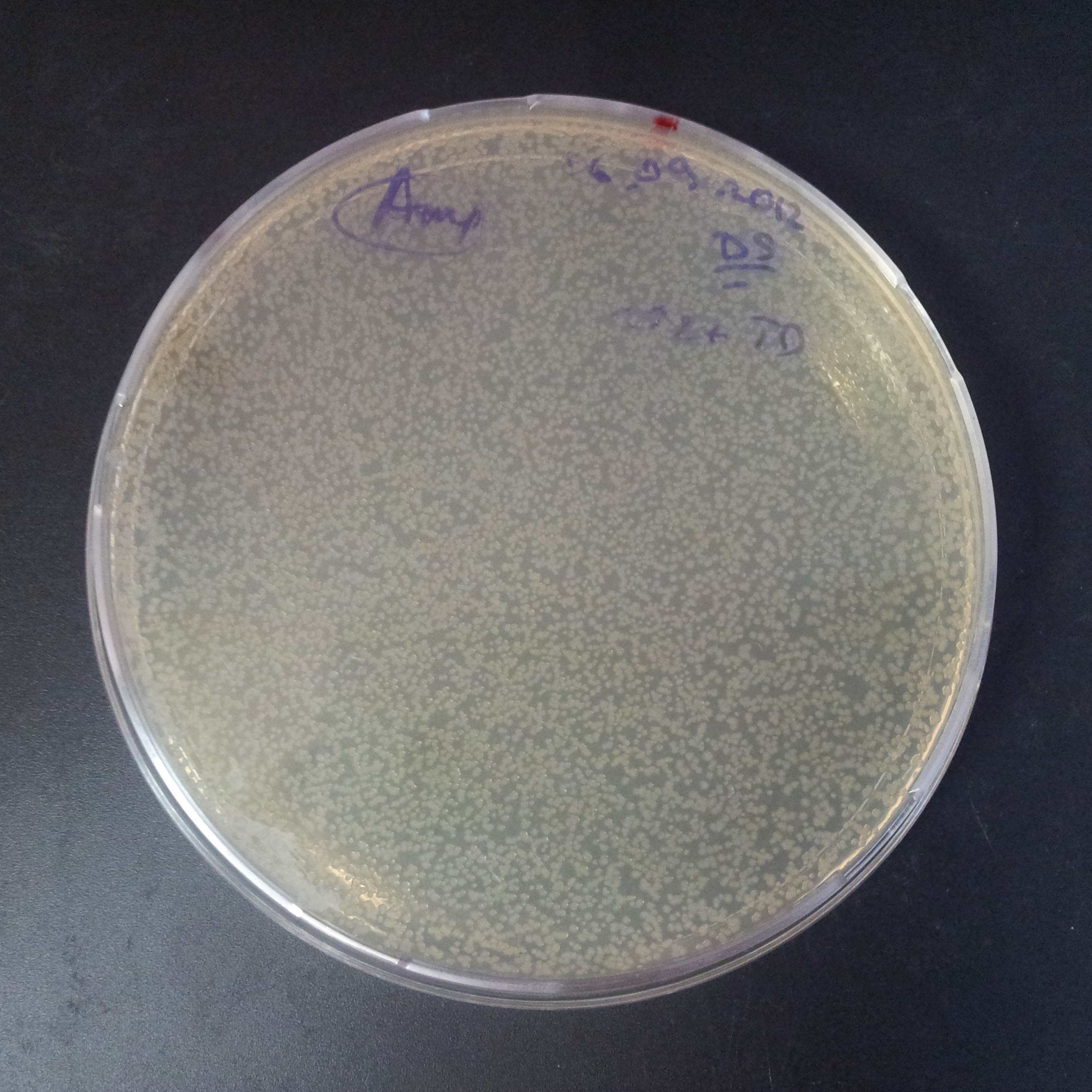
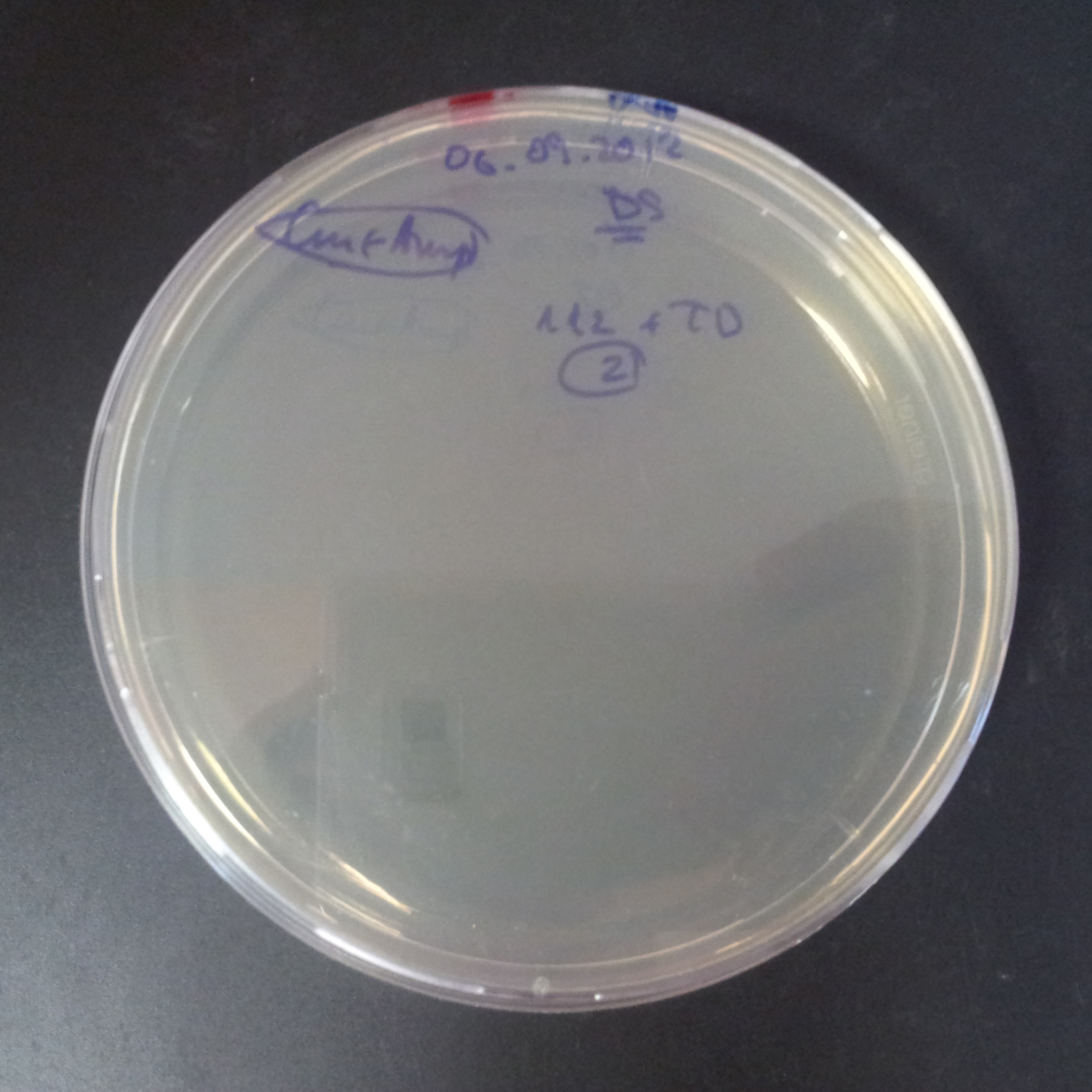
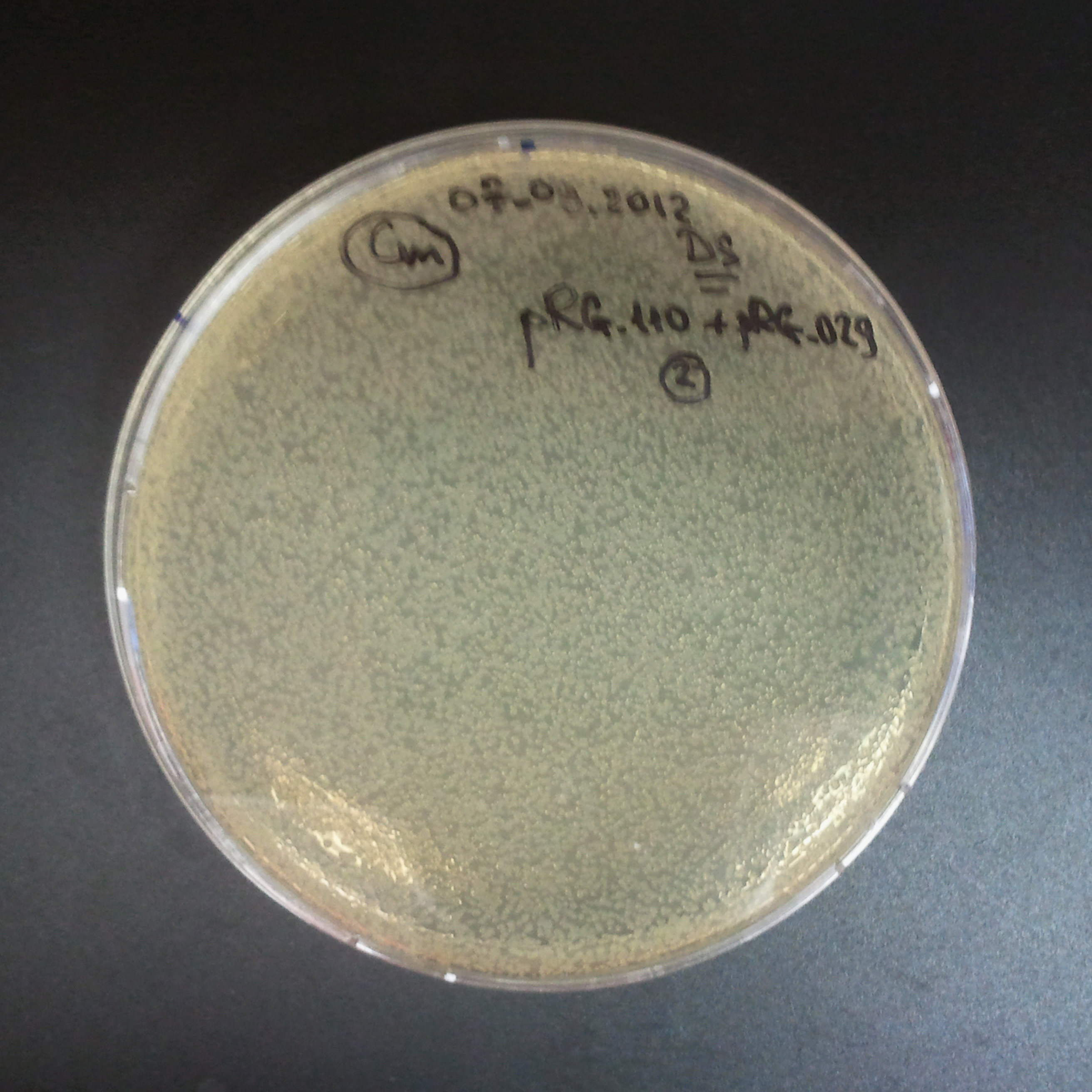
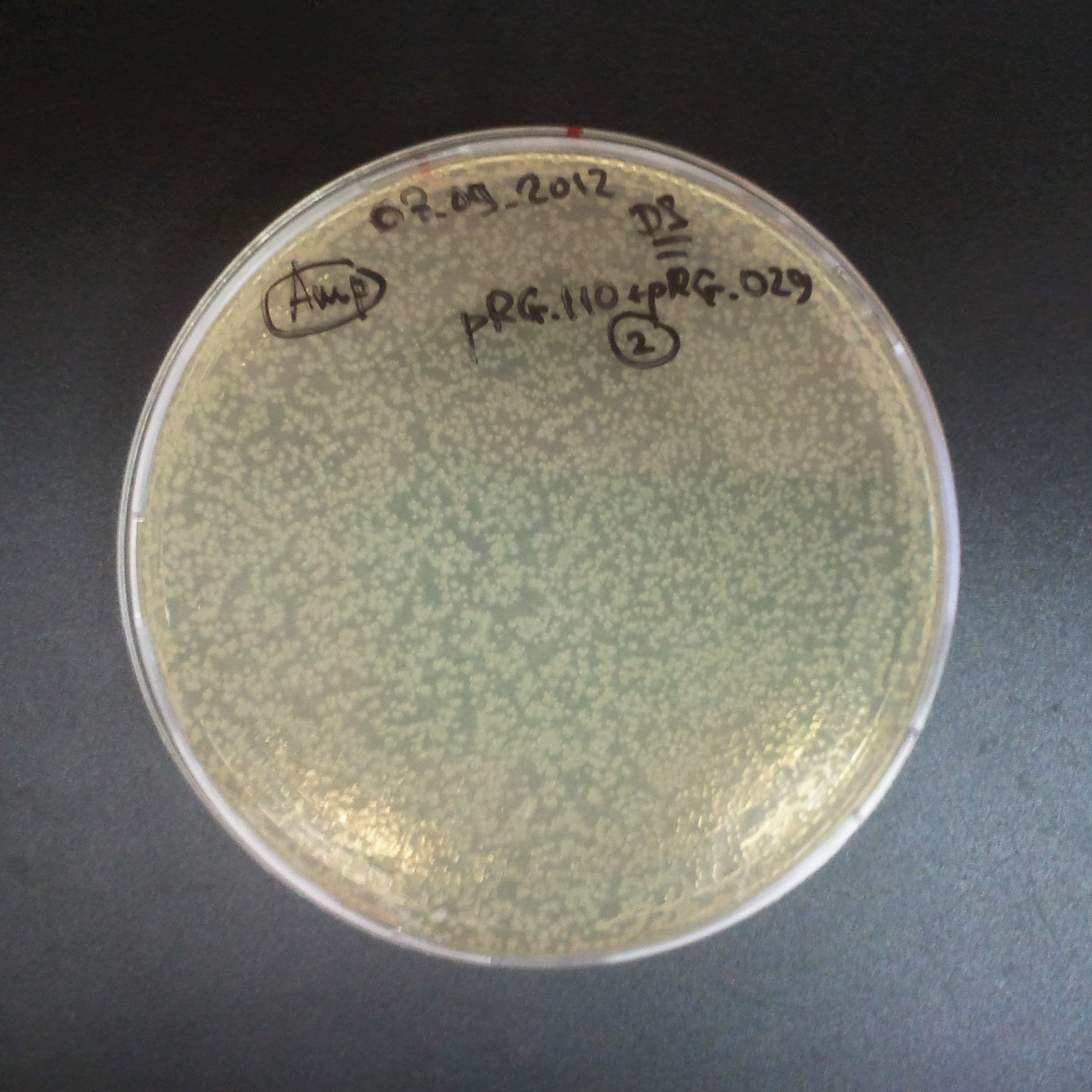
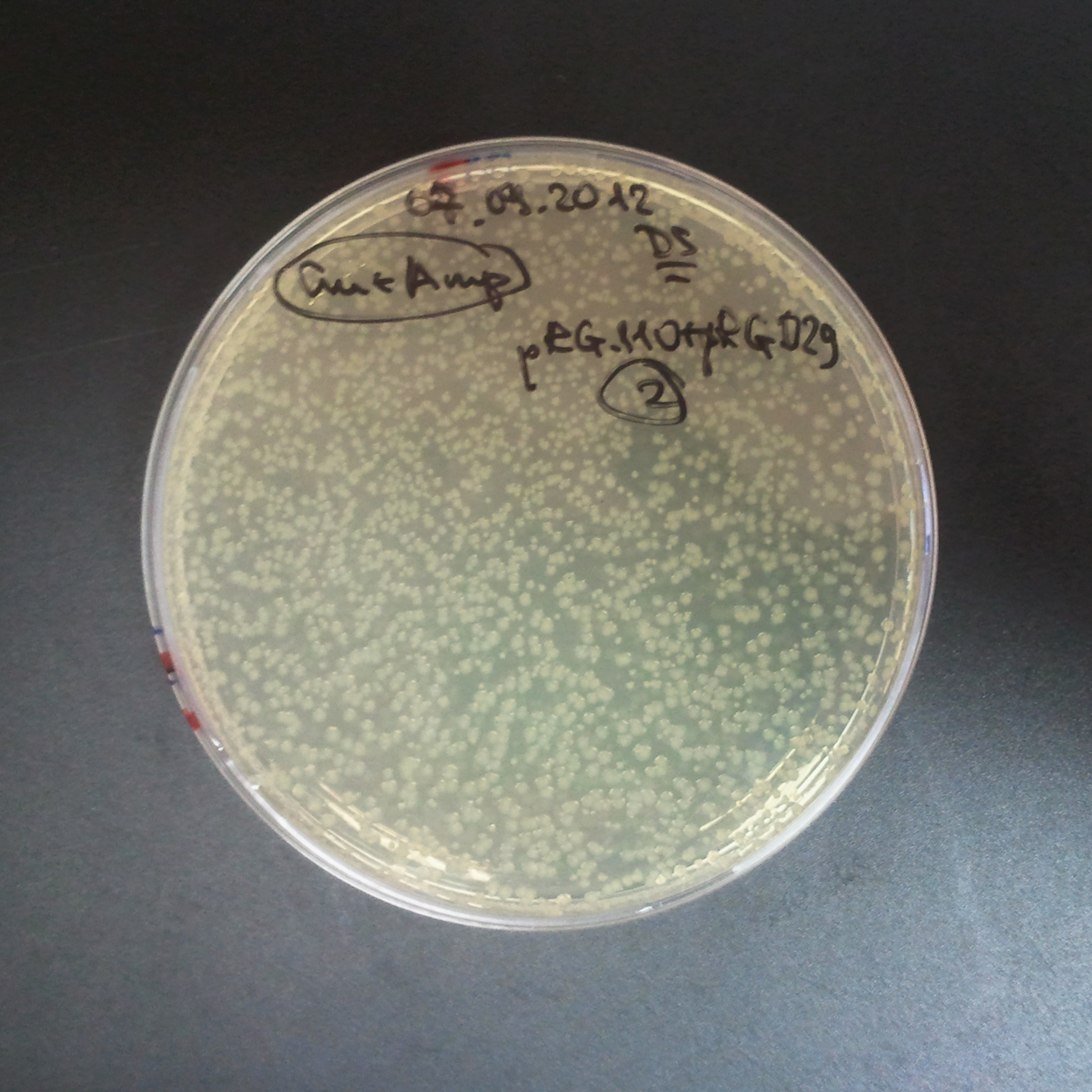
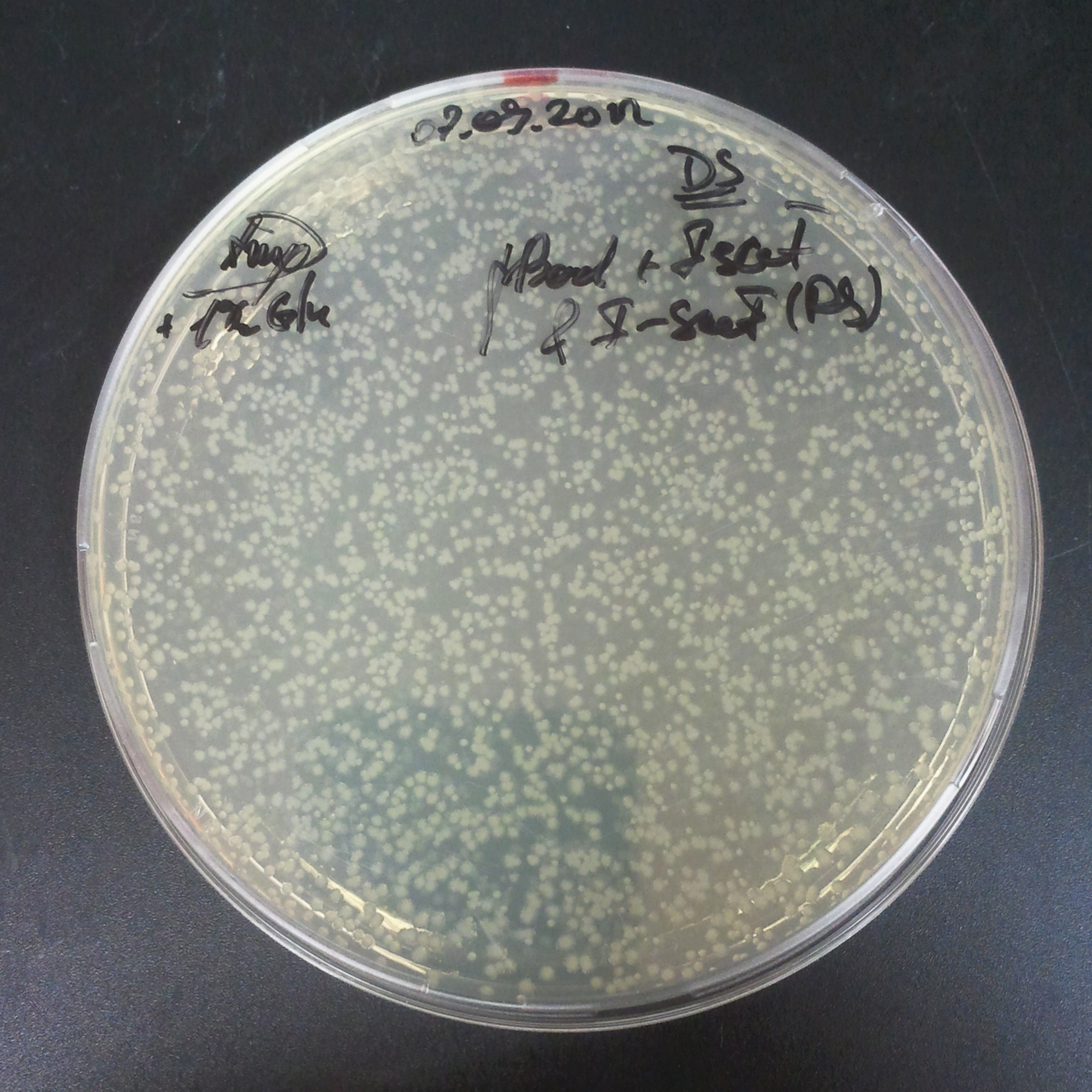
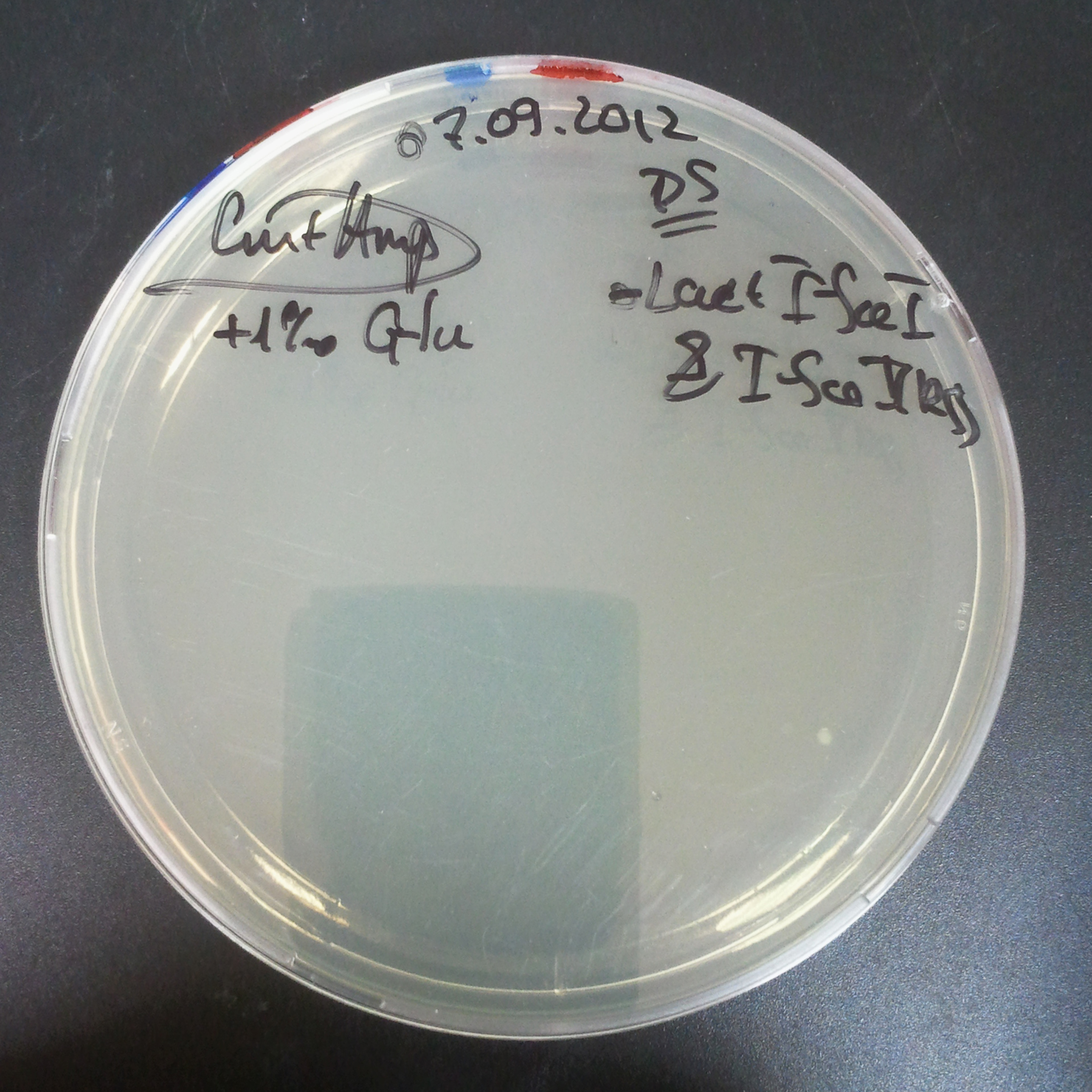
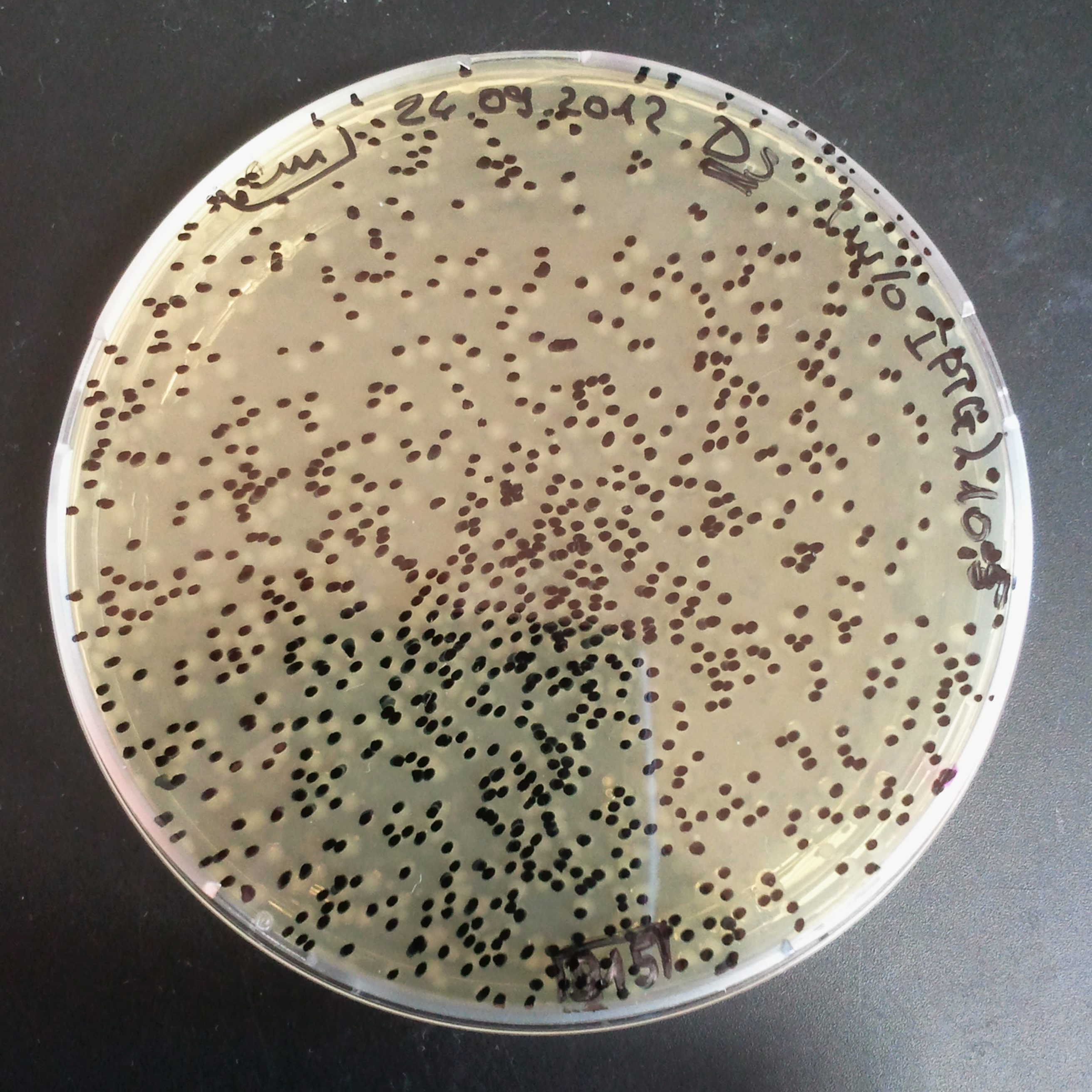
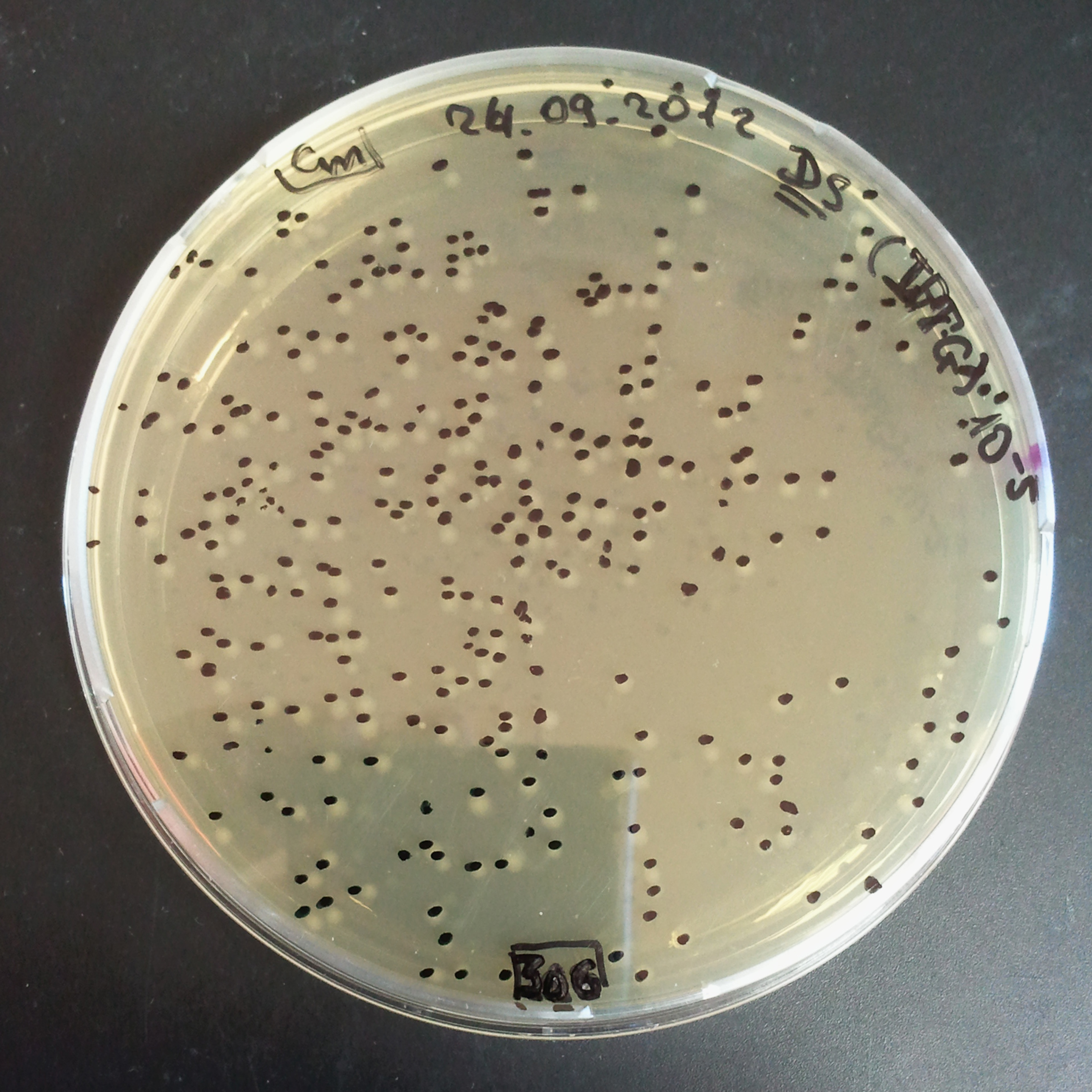
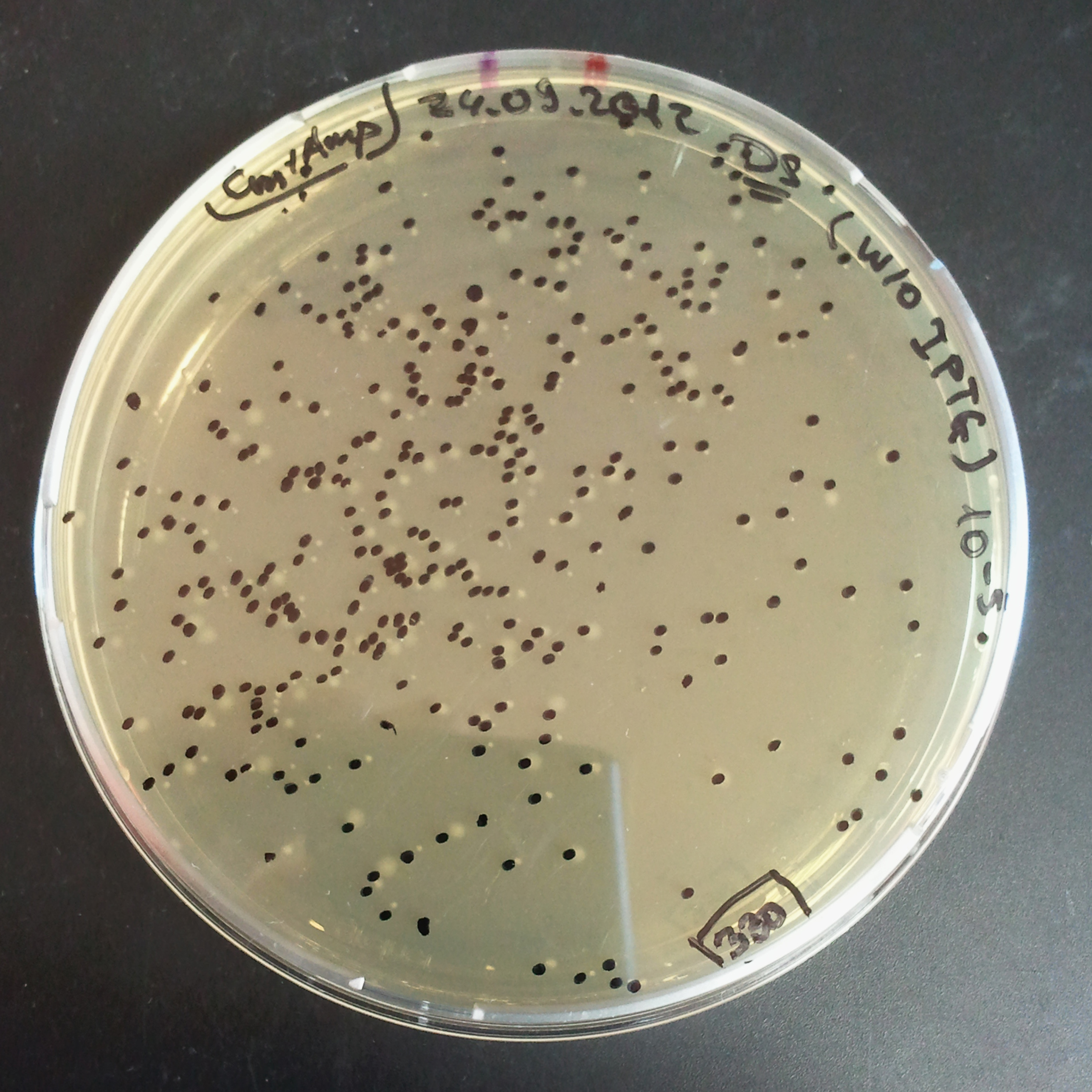
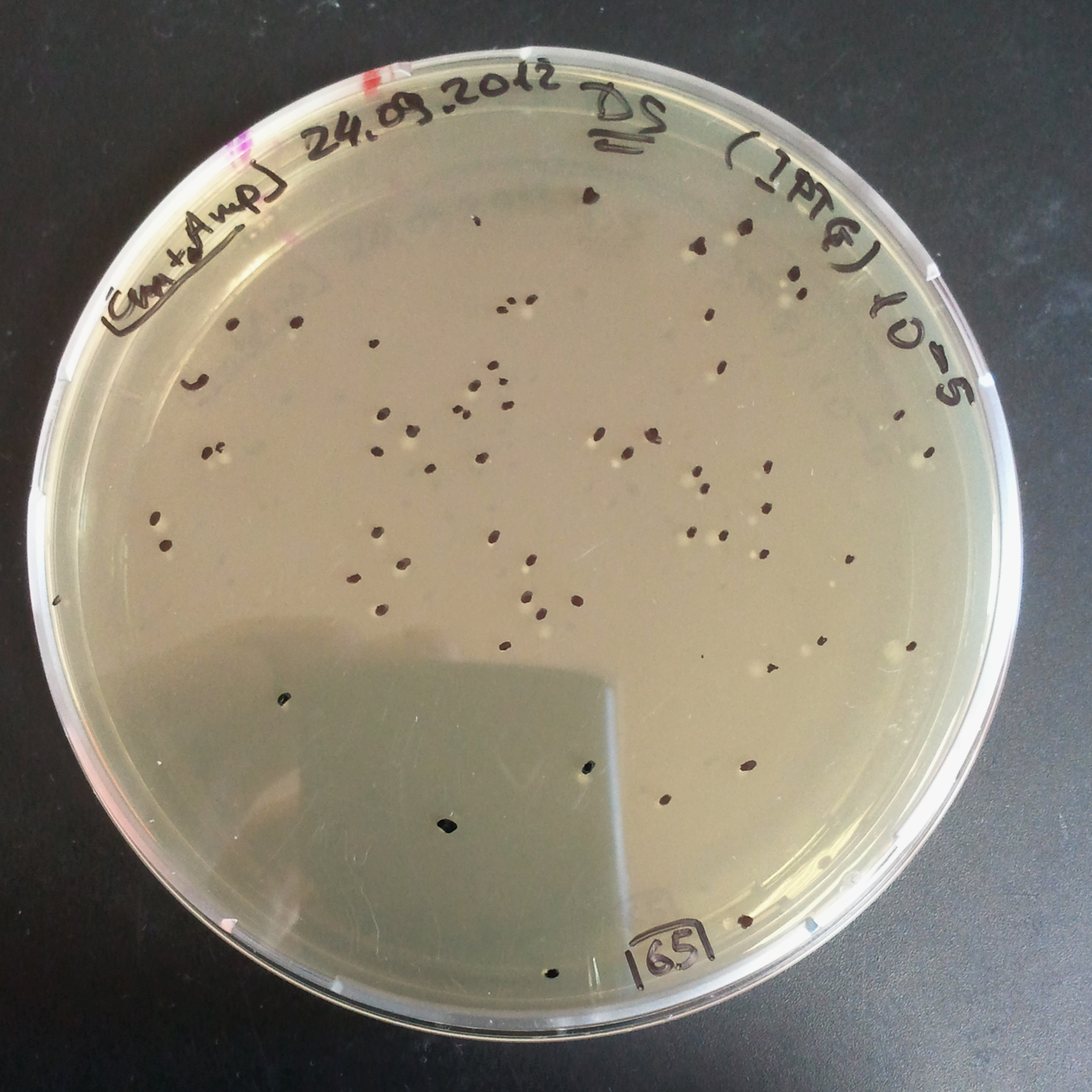
From the experiment we can clearly see that on plates with two antibiotics (Chloramphenicol, Cm; & Ampicillin, Amp) there are no colonies, while on plates with only one antibiotic Cm or Amp there are numerous colonies.
The first combination of plasmids:
- First plasmid: pBad & RBS & I-SceI [Cm]
- Second plasmid: I-SceI restriction site [Amp]
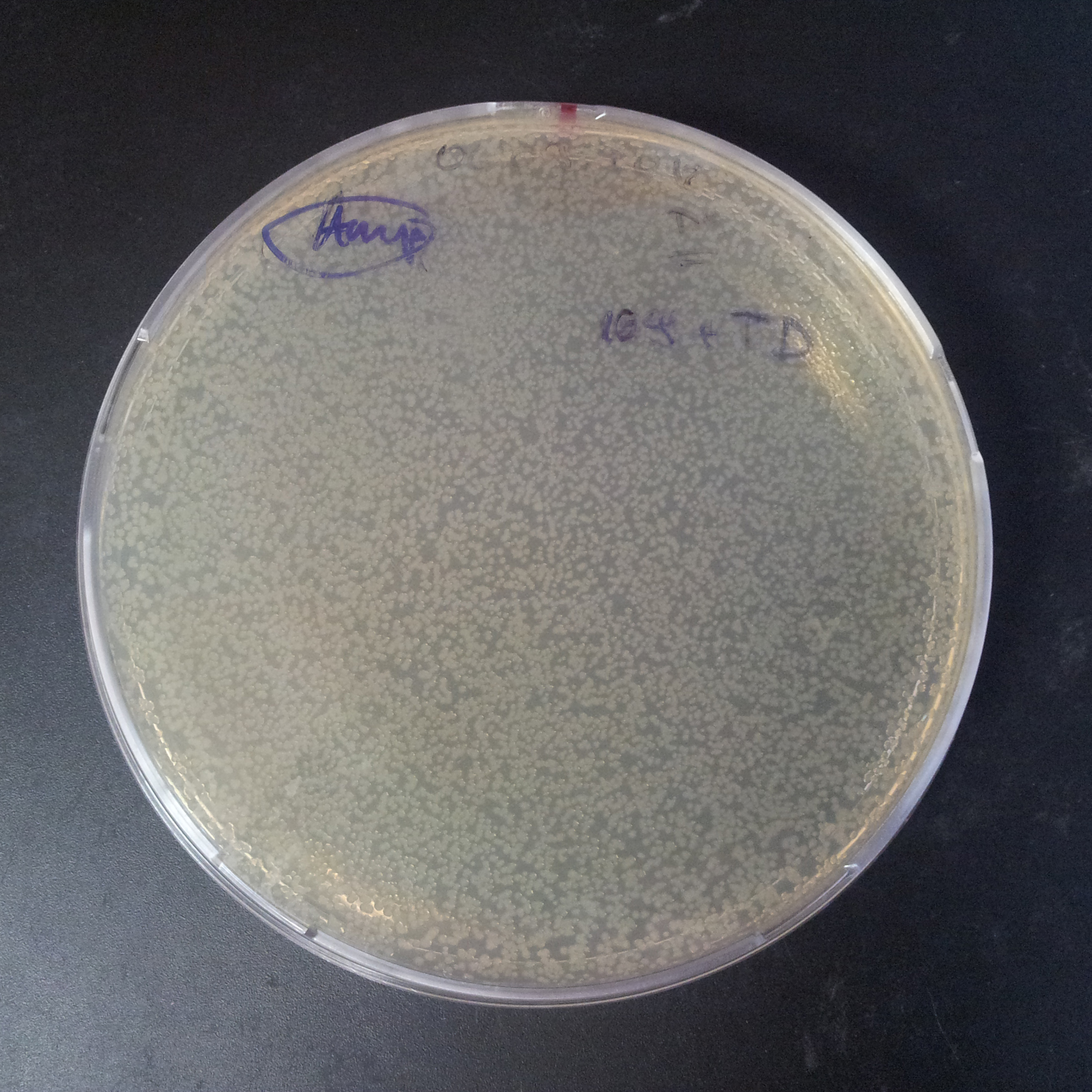
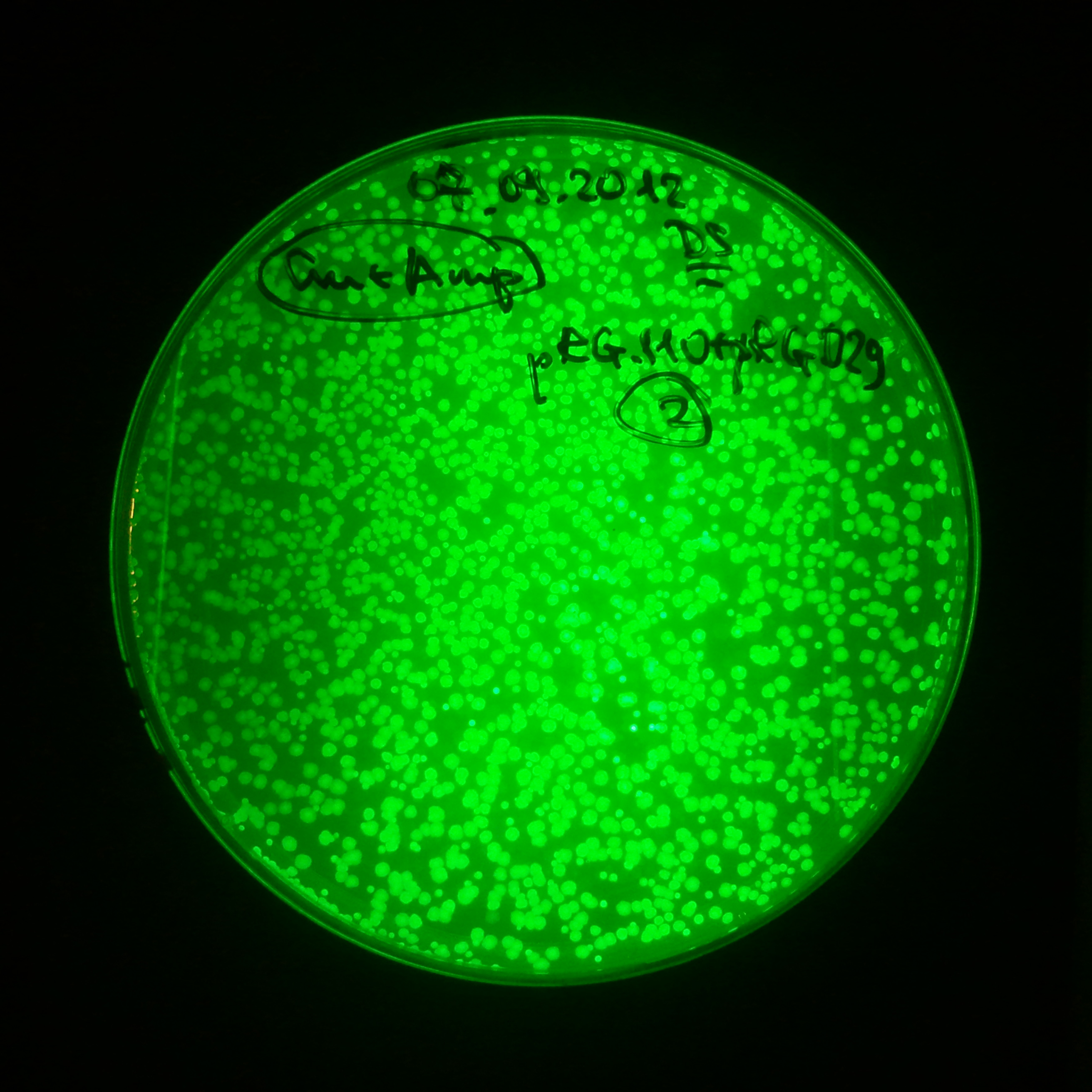
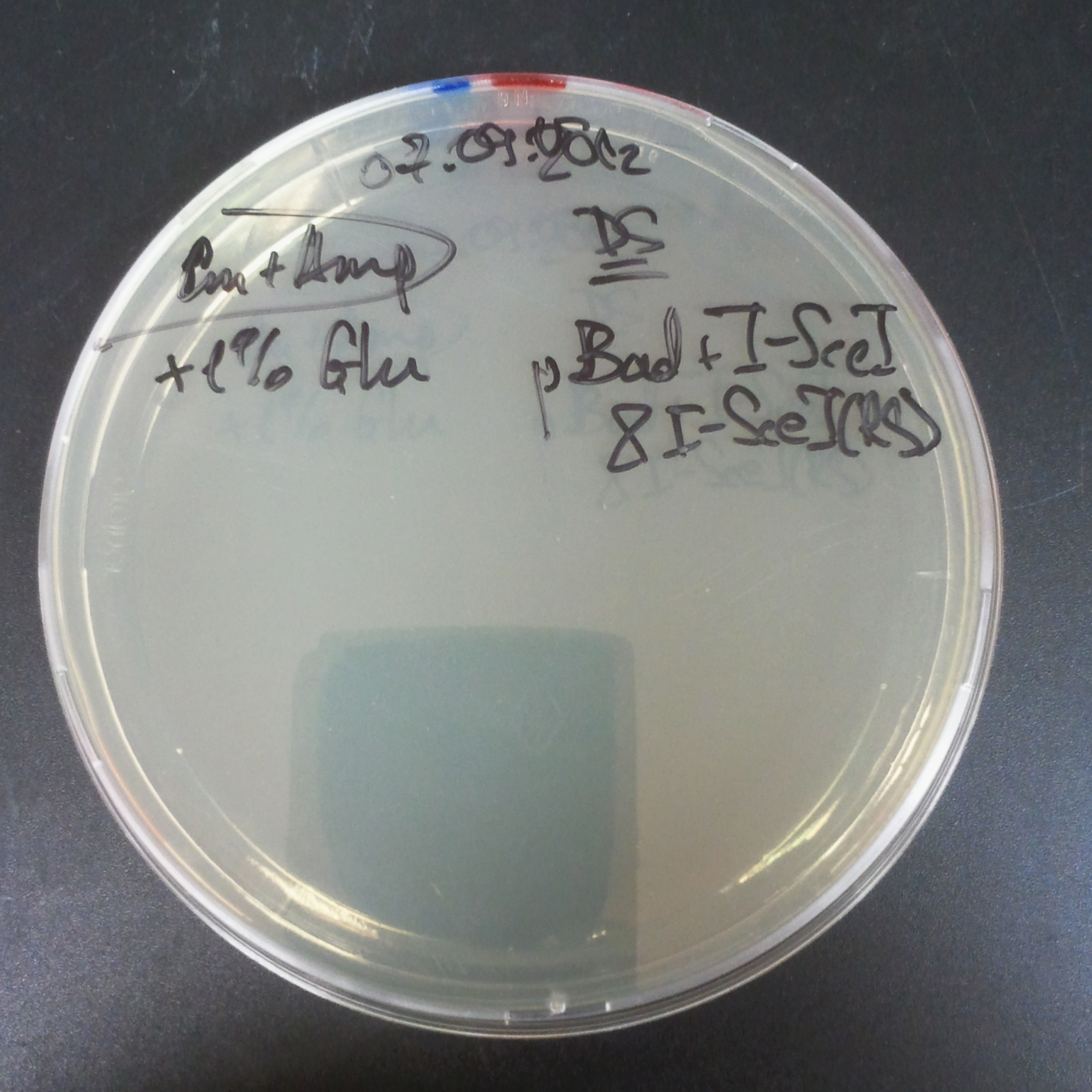
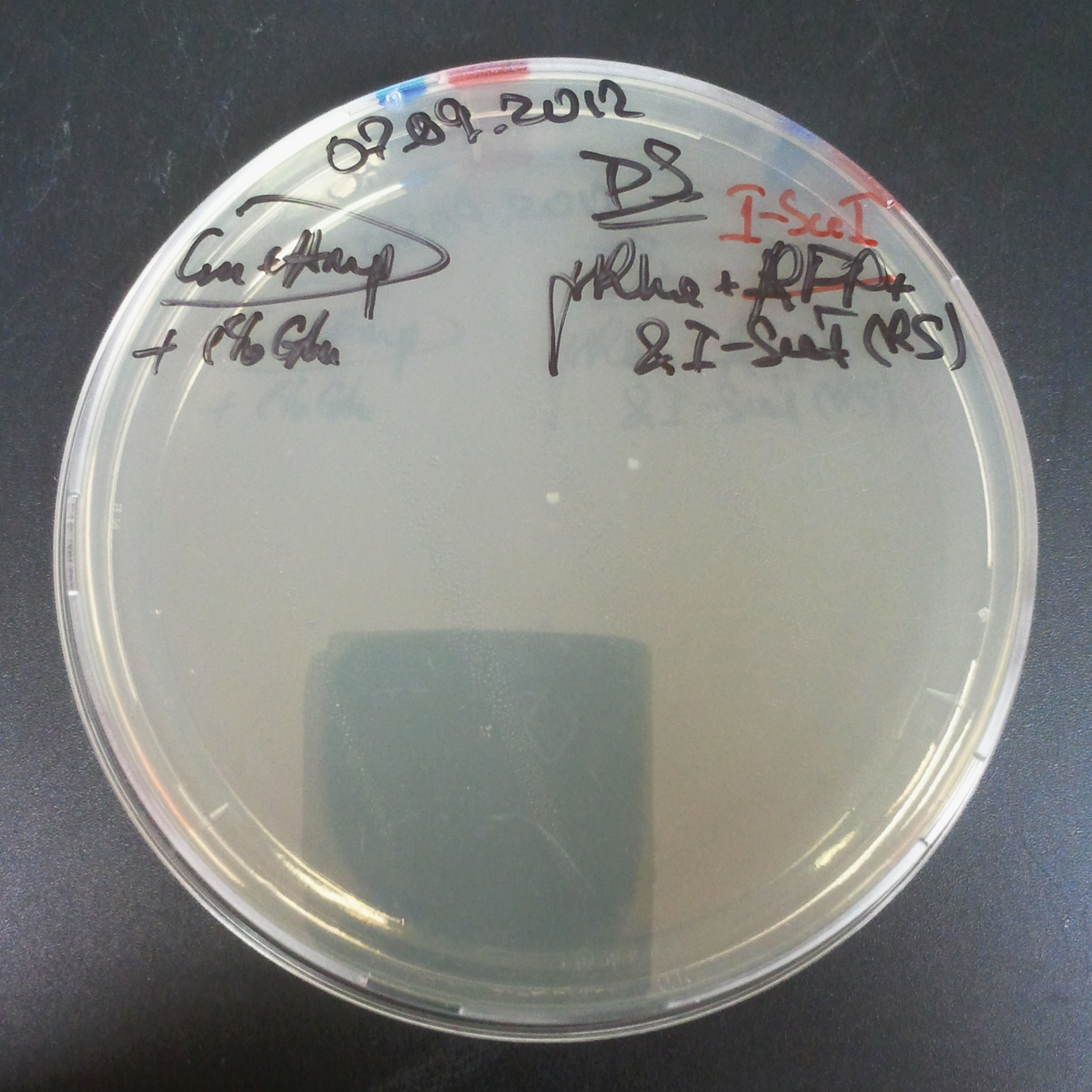
The second combination of plasmids:
- First plasmid: pLac & RBS & I-SceI [Cm]
- Second plasmid: I-SceI restriction site [Amp]
The third combination of plasmids:
- First plasmid: pLac & RBS & I-SceI [Cm]
- Second plasmid: I-SceI restriction site [Amp]
We suggested two hypotheses to explain the results:
- Those two plasmids are not compatible. Plasmids could have different origins of replication. That might be the reason why double transformation is unsuccessful.
- Our system works. Our system perfectly works, but there is some leakage in the promoter leading to the expression of I-SceI meganuclease. In such case, it very efficiently cuts I-SceI restriction site, digesting the second plasmid with ampicillin resistance.
Firstely, we decided to check the first hypothesis, and to check if two plasmids are compatible with each other.
Control for plasmid compatibility
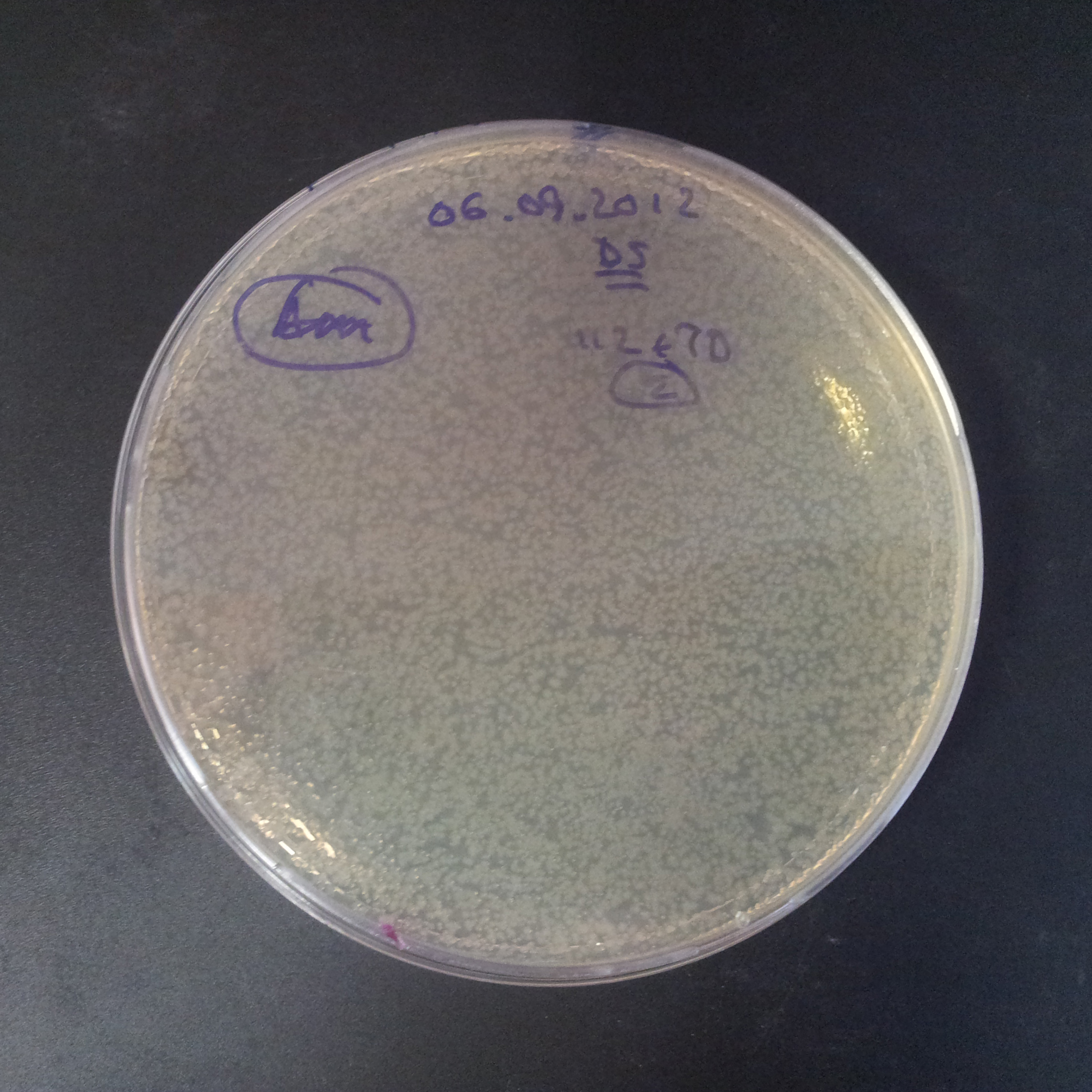
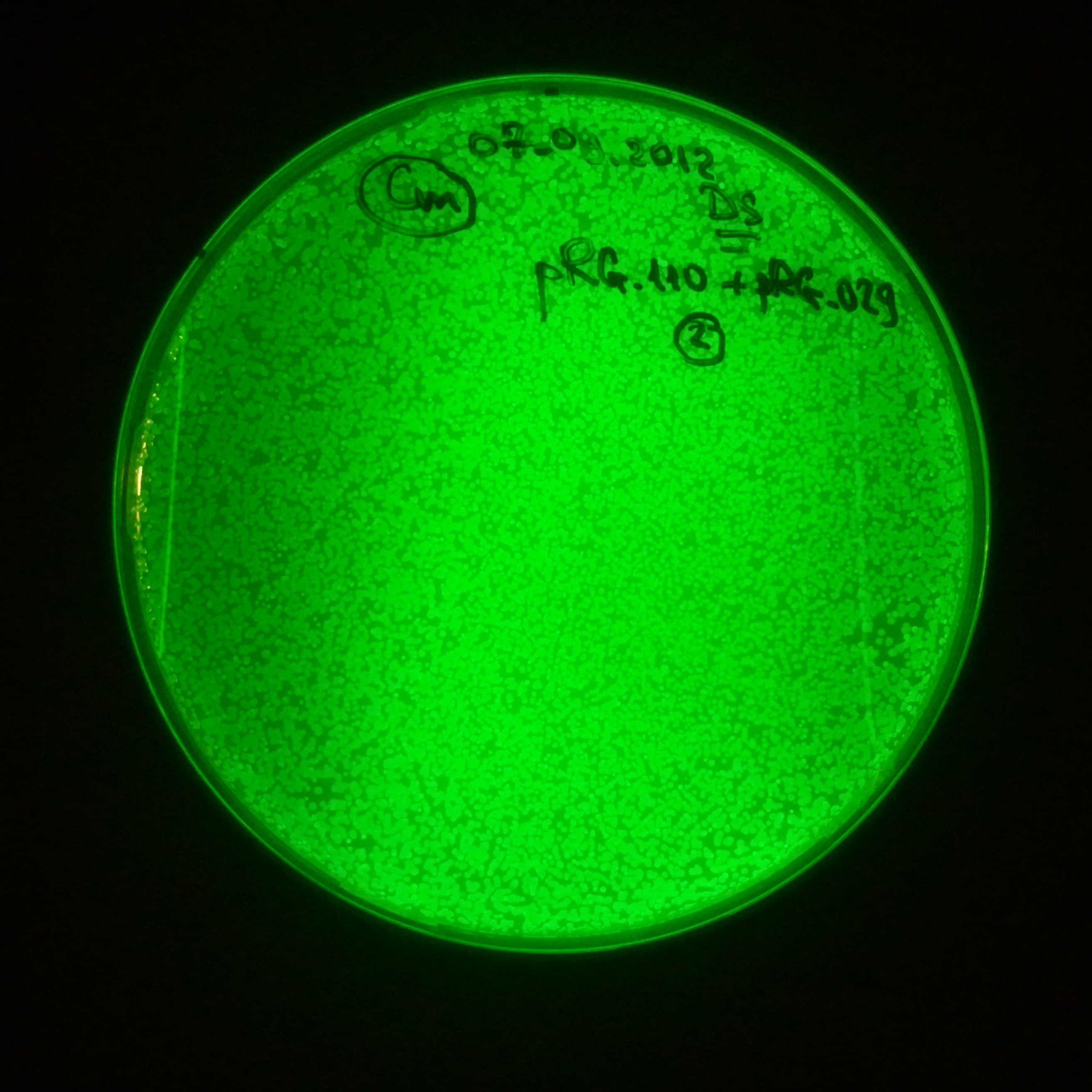
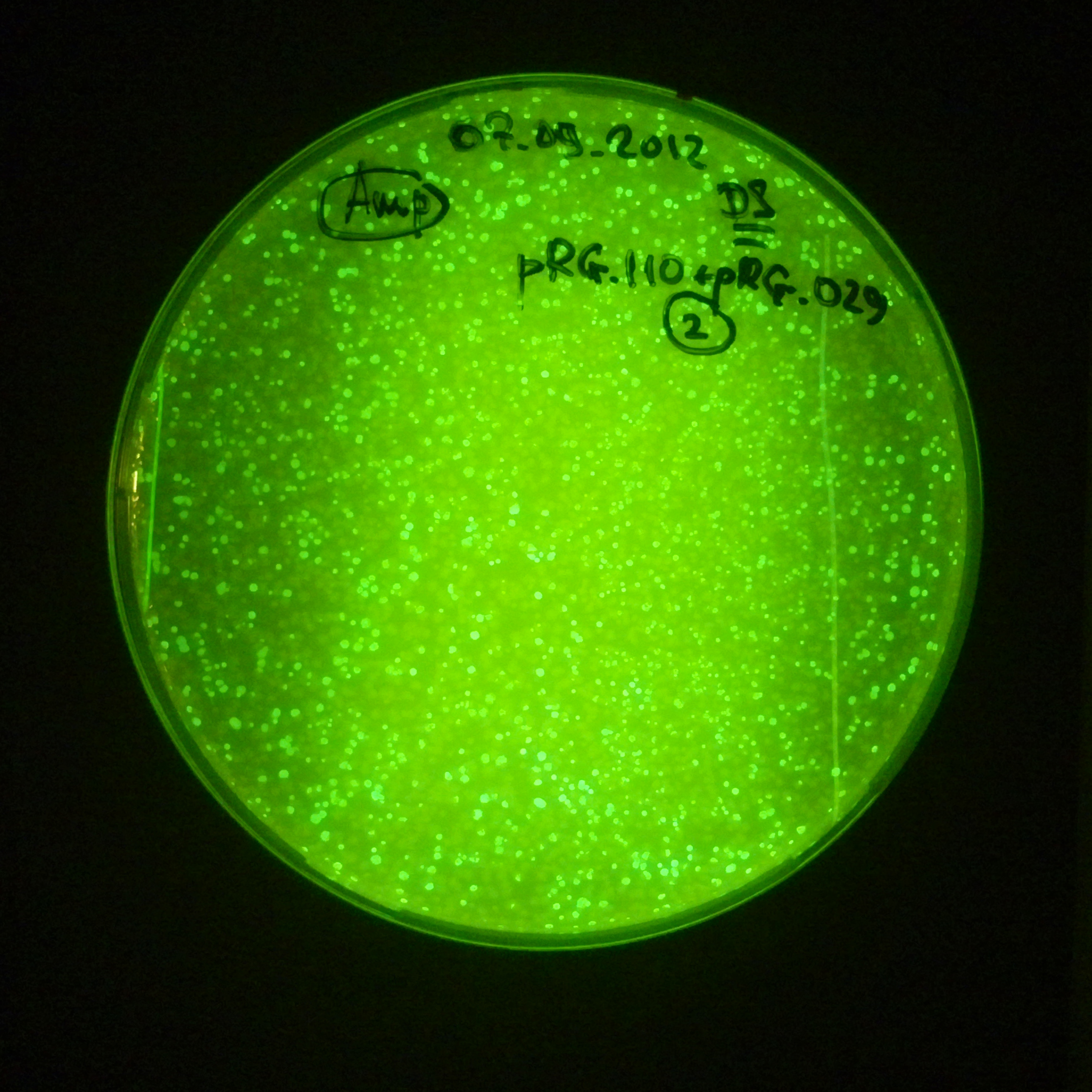
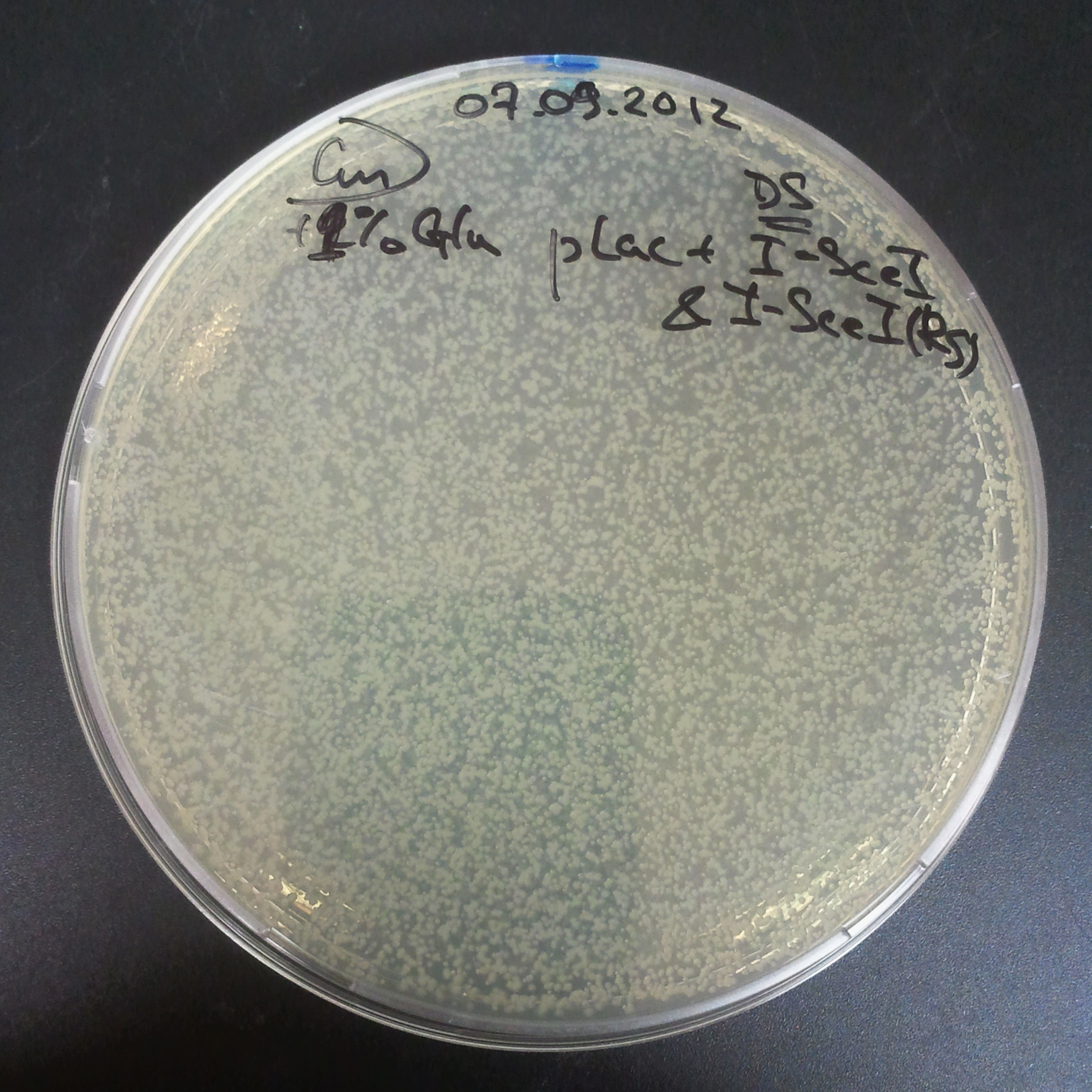
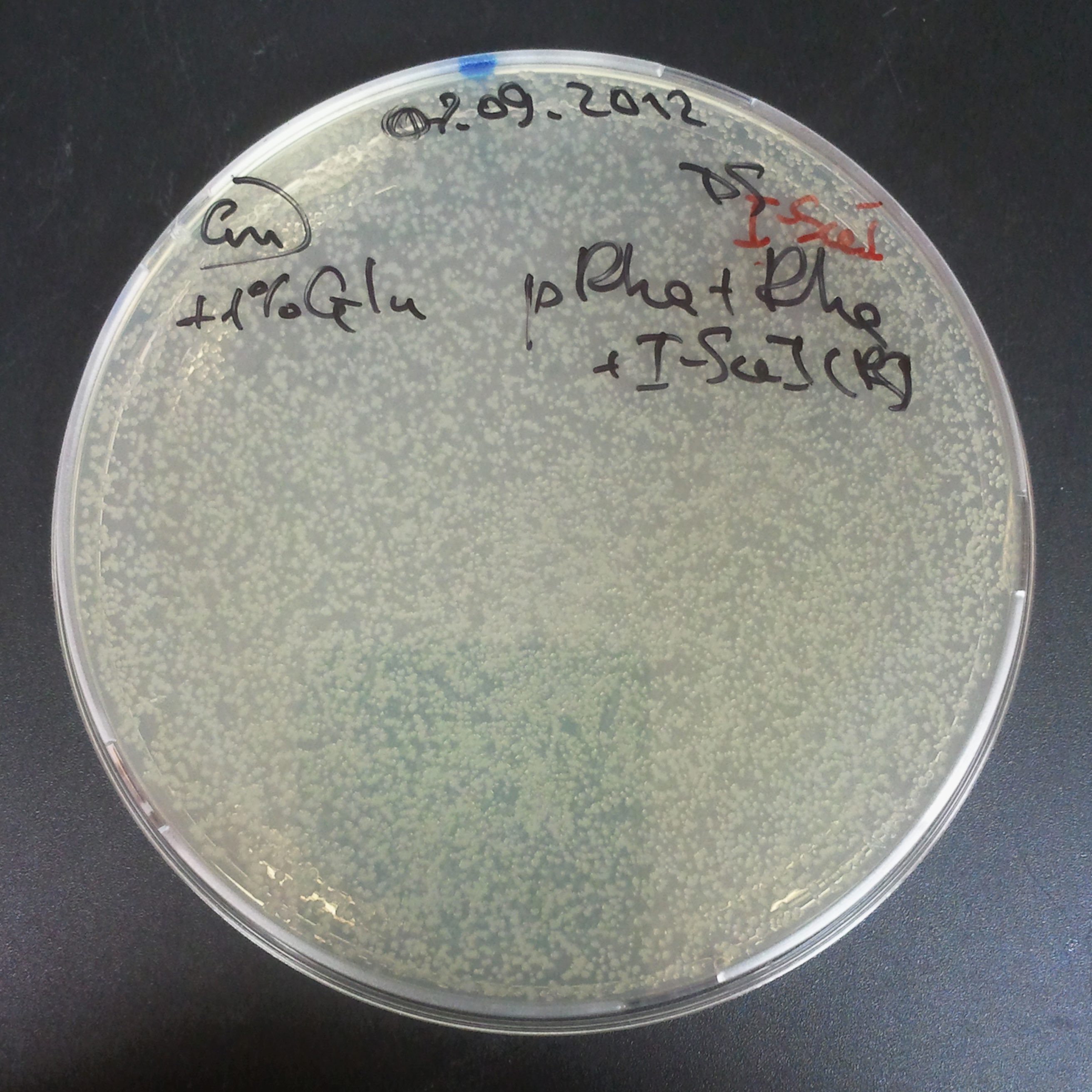
As control experiment, we decided to trasform two plasmids into NEB Turbo E.Coli strain. The first plasmid in this experiment is analogous to the one from the previous experiment, but with GFP insted of I-SceI meganuclease; the second plasmid is the same as in the previous experiment:
- First plasmid: Low copy plasmid with encoded generator to express GFP meganucllease. Only the version with pLac promoter was tested, because they all have the same backbone plasmid, and consequently the same replication prigin.
- Backbone: pSB3C5
- Resistance: Chloramphenicol
- Origin of Replication: modified pMB1 derived from pUC19
- Second plasmid: High copy plasmid with encoded I-SceI restriction site, [http://partsregistry.org/Part:BBa_K175027 K175027].
- Backbone: pSB1AK3
- Resistance: Ampicillin and Kanamycin
- Origin of Replication: modified pMB1 derived from pUC19
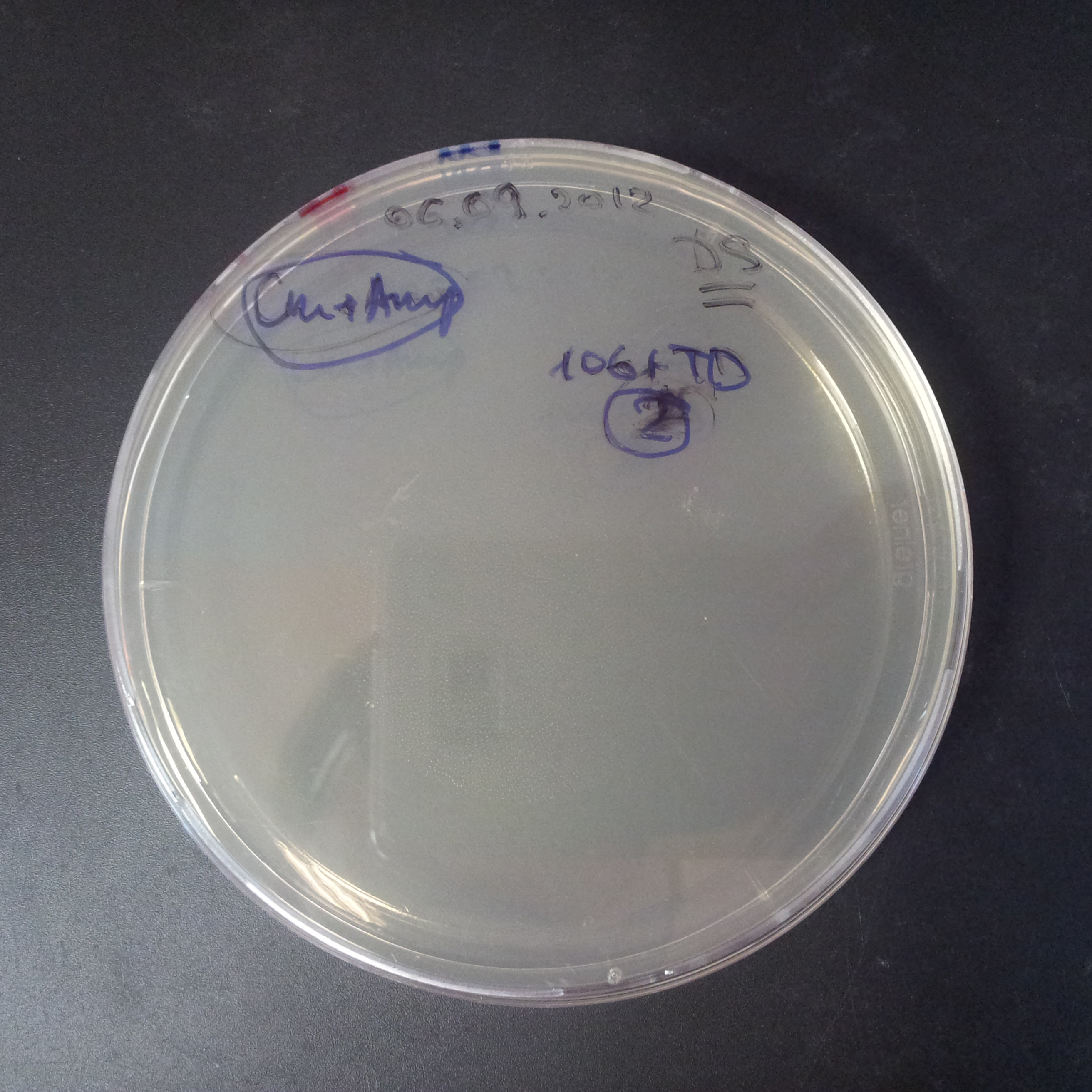
We expected to have colonies on both type of plates: firstly, on plates with one antibiotic (Chloramphenicol & Ampicillin), secondly, on plates with both antibiotics.
Our expectations became true, and our cells expressed GFP, so we conclude these two plasmids have compatible replication origins, and there should be nothing preventing the I-SceI from being expressed in the previous experiment. That means that our circuits work, but there is some leaky expression of I-SceI meganuclease that leads to a very efficient digestion of the plasmid carrying the Ampicillin antibiotic.
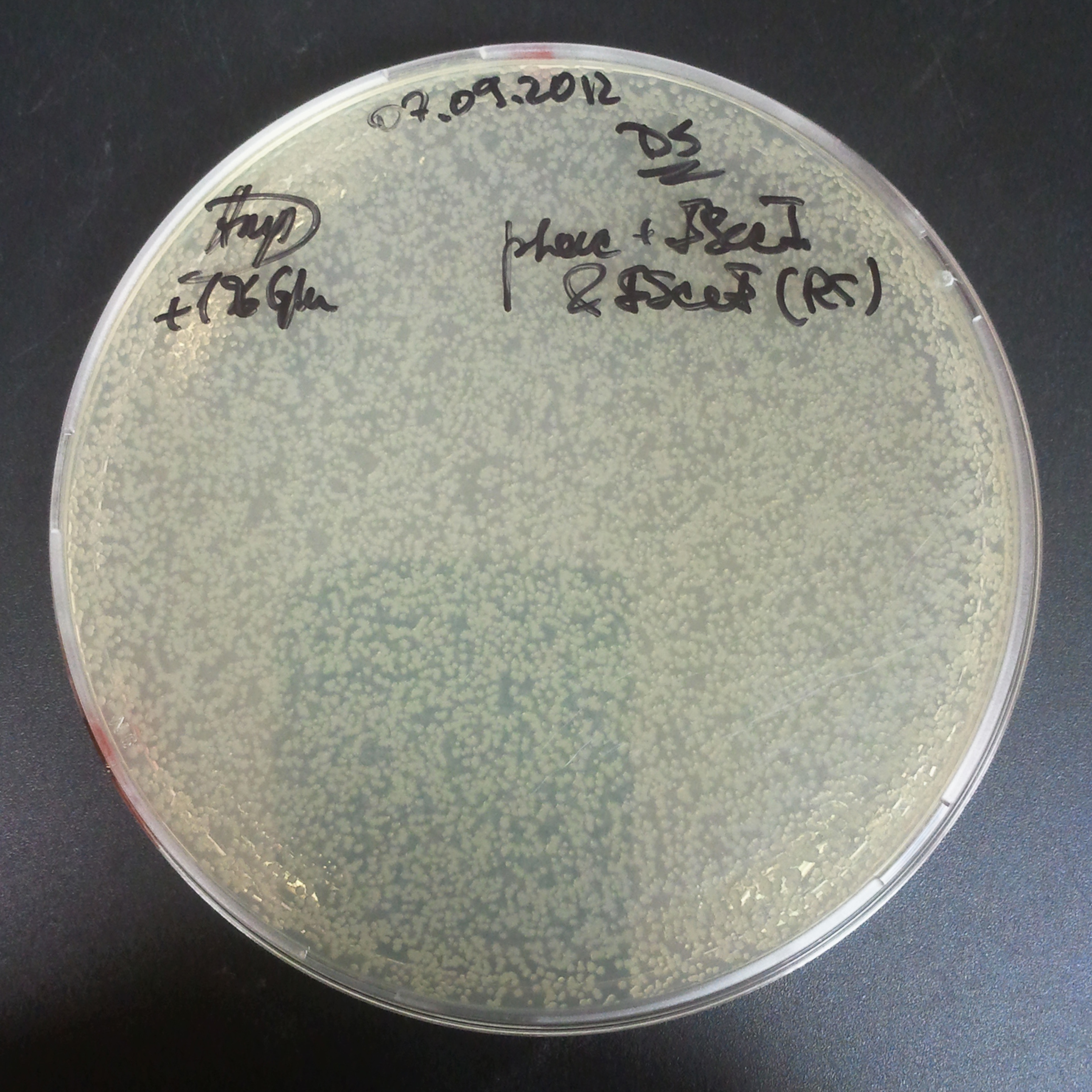
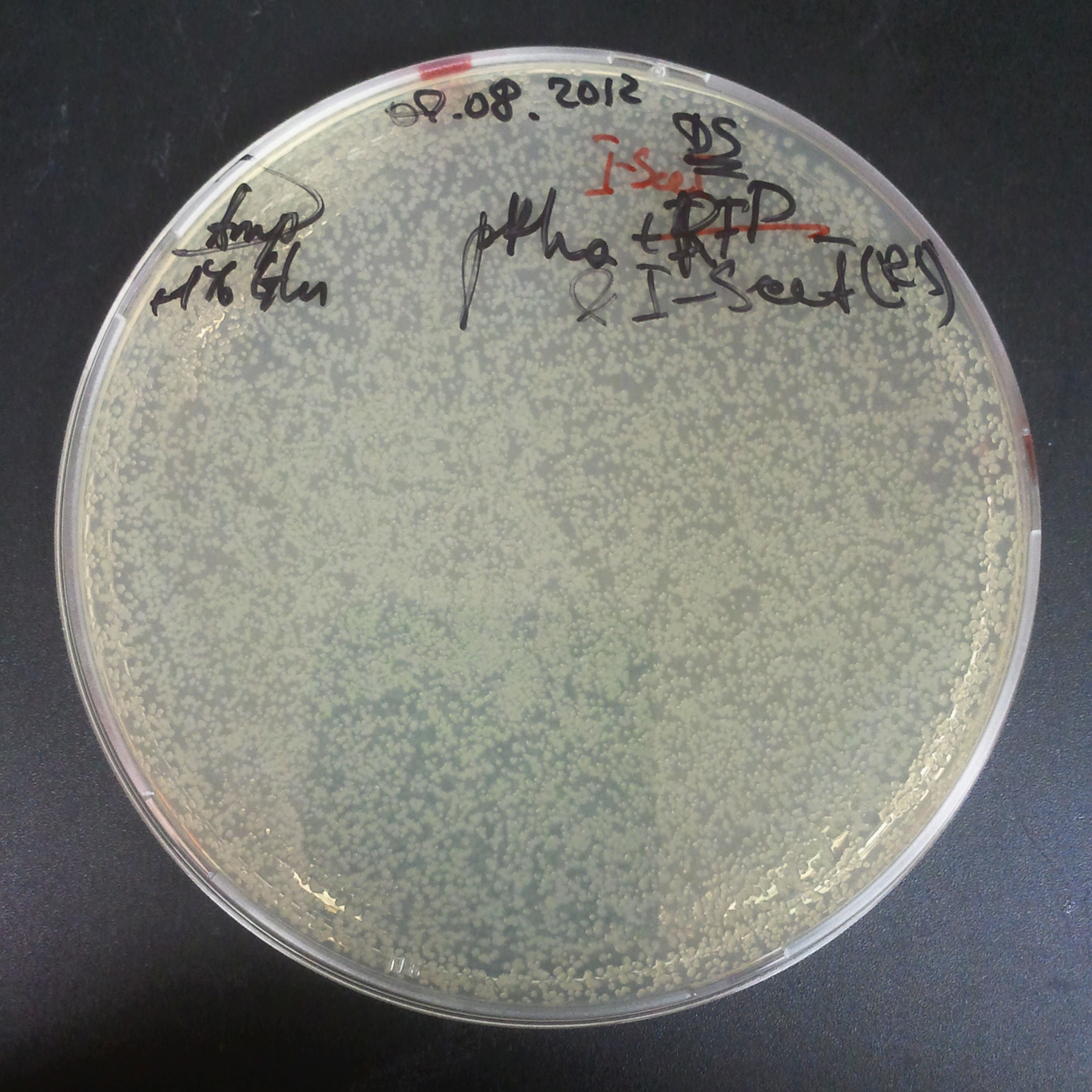
To avoid leakage, in the next experiment we tried to recover cells after transformation and plate it in the presence of glucose that represses the pLac promoter.
Recovery in glucose
Results
Present your results
Measuring the I-SceI efficiency (TUDelft parts)
In 2009, TUDelft iGEM Team has already tried to design a Self Destructive Plasmid based on I-SceI meganuclease. For their experiments, they designed a generator to produce I-SceI meganuclease and used the same pLac promoter, but they had two essential differences with our system:
- They used a [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0030 strong RBS].
- I-SceI meganuclease they used had an LVA tag.
Moreover, they didn't submit the meganuclease alone as a biobrick, so it couldn't be used for constructing new composite biobricks controlled by other promoters, which would be very useful for the modularity of our system. That is why we started working on our own constructions of meganuclease first. However, we asked TUDelft to send us two plasmids that they designed, so that we can test and characterize them:
- First plasmid: High copy plasmid with encoded generator to express I-SceI meganucllease, [http://partsregistry.org/Part:BBa_K175041 BBa_K175041]:
- Backbone: pSB1C3
- Resistance: Chloramphenicol
- Origin of Replication: modified pMB1 derived from pUC19
- Second plasmid: High copy plasmid with encoded I-SceI restriction site, [http://partsregistry.org/Part:BBa_K175027 K175027]:
- Backbone: pSB1AK3
- Resistance: Ampicillin and Kanamycin
- Origin of Replication: modified pMB1 derived from pUC19
Step 1
To mesure the efficiency of I-SceI from TUDelft parts, we proceeded in the same way as for the characterization of our own designs: we transformed two plasmids with different antibiotic resistances into NEB Turbo E.Coli strain.
The transformation was successful.
Step 2
Results
Present your results
Characterisation of pRha
Experimental setup
Describe the experiment
Results
Present your results
References
- «Fsel, a new type II restriction endonuclease that recognizes the octanucleotide sequence 5′ GGCCGGCC 3′», Janise Meyertons Nelson+, Sheila M. Miceli, Mary P. Lechevalier1 and Richard J. Roberts*, (1990)
- «Structure and activity of the mitochondrial intron-encoded endonuclease, I-SceIV», Wernette C. M. Biochem Biophys Res Commun, (1998)
- Yisheng Kang et al., «Systematic Mutagenesis of E.coli K-12 MG1655 ORFs», Yisheng Kang, Tim Durfee, Jeremy D. Glasner, Yu Qiu, David Frisch, Kelly M. Winterberg, and Frederick R. Blattner, (2004)
- «On Spontaneous DNA Damage in Single Living Cells», Jeanine M. Pennington, Ph.D. thesis, Baylor College of Medicine, Houston (2006):
- «A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation», Rebecca G. Ponder, Natalie C. Fonville, Susan M. Rosenberg, (2005)
- «Universal Code Equivalent of a Yeast Mitochondrial lntron Reading Frame Is Expressed into E. coli as a Specific Double Strand Endonuclease», L. Colleaux, L. d'Auriol, M. Betermier, G. Cottarel, A. Jacquier, F. Galibert†, B. Dujon, (1986)
- «Tightly regulated vectors for the cloning and expression of toxic genes», Larry C. Anthony*, Hideki Suzuki, Marcin Filutowicz, (2004)
- «In Vivo Induction Kinetics of the Arabinose Promoters in Escherichia coli» , Casonya M. Johnson And Robert F. Schleif*, (1995)
- «Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system» Matthew J. Giacalone1, Angela M. Gentile2, Brian T. Lovitt2, Neil L. Berkley2, Carl W. Gunderson1, and Mark W. Surber, (2006)
- «DNA-Dependent Renaturation of an insoluble DNA binding Protein. Identification of the RhaS Binding Site at rhaBAD» Susan M.Egan and Robert F. Schleif, (1994)
- «Optimization of an E. coli L-rhamnose-inducible expression vector: test of various genetic module combinations» Angelika Wegerer, Tianqi Sun and Josef Altenbuchner, (2008)
 "
"


 Overview
Overview Delay system
Delay system Semantic containment
Semantic containment Restriction enzyme system
Restriction enzyme system MAGE
MAGE Encapsulation
Encapsulation Synthetic import domain
Synthetic import domain Safety Questions
Safety Questions Safety Assessment
Safety Assessment