Team:University College London/Module 2/Modelling
From 2012.igem.org
Erinoerton (Talk | contribs) (→Modelling) |
Erinoerton (Talk | contribs) (→Modelling) |
||
| Line 9: | Line 9: | ||
[[File:aggnet.png]] | [[File:aggnet.png]] | ||
| - | |||
| + | ==Species == | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Species!! Initial value (molecules) !! Notes | ||
| + | |- | ||
| + | | PE|| 0.044 || Polyethylene found in North Pacific Gyre (value per cubic metre)<sup>1,2</sup> | ||
| + | |- | ||
| + | | POPex || 0.0 || Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface | ||
| + | |- | ||
| + | | PEPOPex || 9.24E-5 || Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface<sup>3</sup> | ||
| + | |- | ||
| + | | POPin || 0.5 || Persistent organic pollutants (in = intracellular) assumed from <i>E. coli</i> membrane permeability <sup>4</sup> | ||
| + | |- | ||
| + | | mRNANahR || 0.0|| NahR mRNA product | ||
| + | |- | ||
| + | | POPinNahR || 0.0|| Complex of the above two molecules | ||
| + | |- | ||
| + | | POPinNahRpSal || 0.0 || Complex of the above molecule and pSal (promoter that induces laccase transcription) | ||
| + | |- | ||
| + | | mRNACurli || 0.0 || Polycistronic mRNA as it codes for more than one protein, in reality curli cluster contains five or more proteins, in our model mRNA is present as ??CsgE??<sup>5</sup> | ||
| + | |- | ||
| + | | CsgA || 0.0 || One of the polypeptides that is coded for in curli cluster, it is a structural component secreted in subunits outside of the cell | ||
| + | |- | ||
| + | | CsgB || 0.0 || One of the polypeptides that is coded for in curli cluster, it is secreted outside of the cell allowing polymerization of CsgA | ||
| + | |- | ||
| + | | Fibril || Result of CsgA and CsgB interaction | ||
| + | |} | ||
| + | |||
| + | ==Reactions taking place in the model== | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Number !! Reaction !! Reaction rate (molecules/sec) !! Notes | ||
| + | |- | ||
| + | | R1 || PE + POPex ↔ PEPOPex || Forward: 1000 <br /> Backward: 1 || Pops have 1000 to 10000 times greater tendency to adhere to plastic than float free in the ocean<sup>6</sup> | ||
| + | |- | ||
| + | | R2 || POPex ↔ POPin || Forward: 0.6 <br /> Backward: 0.4 || Based on membrane permeability<sup>4</sup>: diffusion gradient | ||
| + | |- | ||
| + | | R3 || POPin + mRNA.Nahr → POPin.mRNA.Nahr || Forward: 1 <br /> Backward: 0.0001 || Based on the assumption that the chemical structure/size of POPs is similar to salycilate<sup>6</sup>. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. | ||
| + | |- | ||
| + | | R7 || 0 → mRNA.Nahr || Forward: 0.088 <br /> Backward: 0.6 || Transcription rate of NahR in molecules/sec (for NahR size 909 bp<sup>7</sup>, transcription rate in E.coli 80bp/sec<sup>8</sup>) under constitutive promoter control | ||
| + | |- | ||
| + | | R4 || POPinmRNANahr → POPinmRNANahr.Psal || Forward: 78200 <br /> Backward: 0.191 <sup>9</sup> || NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding | ||
| + | |- | ||
| + | | R5 || POPexmRNANahr.Psal → GFP.mRNA|| 0.11 || Transcription rate of GFP in molecules/sec (for GFP size 720bp<sup>10</sup>, transcription rate in E.coli 80bp/sec<sup>8</sup>) | ||
| + | |- | ||
| + | | R6 || GFP.mRNA → GFP|| 0.084 || Translation rate of GFP in molecules/sec (for GFP size 240aa<sup>10</sup>, translation rate in E.coli 20aa/sec<sup>8</sup>) | ||
| + | |- | ||
| + | | R9 || GFP.mRNA → 0 || 0.03 || Degradation rate of GFP mRNA product<sup>11</sup> must be taken into account due to suboptimal conditions | ||
| + | |} | ||
| + | |||
| + | == Results == | ||
| + | |||
| + | To follow | ||
| + | |||
| + | == References == | ||
| + | 1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect. <i>Biology Letters</i> 10.1098 | ||
| + | |||
| + | 2. Andrady AL (2011) Microplastics in the marine environment. Marine Pollution Bulletin 62: 1596-1605 | ||
Shala AA, Restrepo S, Gonzalez Barrios AF (2011) A network model for biofilm development in Escherichia coli K-12. <i> Theoretical Biology and Medical Modelling </i> 8: 34 doi:10.1186/1742-4682-8-34 | Shala AA, Restrepo S, Gonzalez Barrios AF (2011) A network model for biofilm development in Escherichia coli K-12. <i> Theoretical Biology and Medical Modelling </i> 8: 34 doi:10.1186/1742-4682-8-34 | ||
| + | |||
| + | 3. To follow | ||
| + | |||
| + | 4. To follow | ||
| + | |||
| + | 5. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. <i>Mol Microbiol</i> 81: 486-499 | ||
| + | |||
| + | 6. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324 | ||
| + | |||
Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB (1997) Amyloid b-Protein Fibrillogenesis: Detection of a protofibrillar intermediate. <i> The Journal of Biological Chemistry </i> 272: 22364–22372 | Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB (1997) Amyloid b-Protein Fibrillogenesis: Detection of a protofibrillar intermediate. <i> The Journal of Biological Chemistry </i> 272: 22364–22372 | ||
Revision as of 16:30, 21 September 2012
Contents |
Module 2: Aggregation
Description | Design | Construction | Characterisation | Shear Device | Modelling | Results | Conclusions
Modelling
In addition to our gene network model for this module, we have come up with a system of equations which describe the strength of our adhesive biofilms under shear force stress.
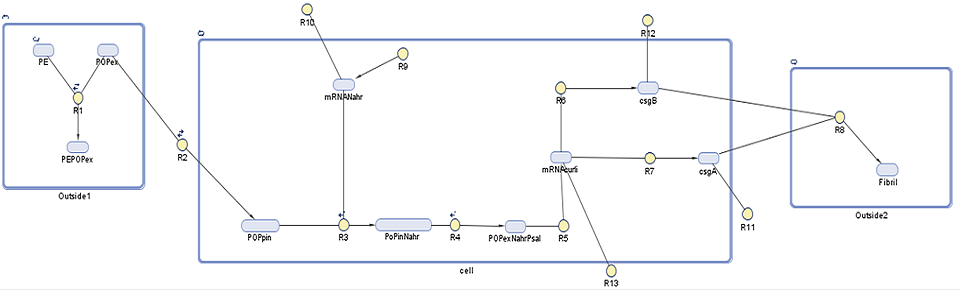
Species
| Species | Initial value (molecules) | Notes |
|---|---|---|
| PE | 0.044 | Polyethylene found in North Pacific Gyre (value per cubic metre)1,2 |
| POPex | 0.0 | Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface |
| PEPOPex | 9.24E-5 | Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface3 |
| POPin | 0.5 | Persistent organic pollutants (in = intracellular) assumed from E. coli membrane permeability 4 |
| mRNANahR | 0.0 | NahR mRNA product |
| POPinNahR | 0.0 | Complex of the above two molecules |
| POPinNahRpSal | 0.0 | Complex of the above molecule and pSal (promoter that induces laccase transcription) |
| mRNACurli | 0.0 | Polycistronic mRNA as it codes for more than one protein, in reality curli cluster contains five or more proteins, in our model mRNA is present as ??CsgE??5 |
| CsgA | 0.0 | One of the polypeptides that is coded for in curli cluster, it is a structural component secreted in subunits outside of the cell |
| CsgB | 0.0 | One of the polypeptides that is coded for in curli cluster, it is secreted outside of the cell allowing polymerization of CsgA |
| Fibril | Result of CsgA and CsgB interaction |
Reactions taking place in the model
| Number | Reaction | Reaction rate (molecules/sec) | Notes |
|---|---|---|---|
| R1 | PE + POPex ↔ PEPOPex | Forward: 1000 Backward: 1 | Pops have 1000 to 10000 times greater tendency to adhere to plastic than float free in the ocean6 |
| R2 | POPex ↔ POPin | Forward: 0.6 Backward: 0.4 | Based on membrane permeability4: diffusion gradient |
| R3 | POPin + mRNA.Nahr → POPin.mRNA.Nahr | Forward: 1 Backward: 0.0001 | Based on the assumption that the chemical structure/size of POPs is similar to salycilate6. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. |
| R7 | 0 → mRNA.Nahr | Forward: 0.088 Backward: 0.6 | Transcription rate of NahR in molecules/sec (for NahR size 909 bp7, transcription rate in E.coli 80bp/sec8) under constitutive promoter control |
| R4 | POPinmRNANahr → POPinmRNANahr.Psal | Forward: 78200 Backward: 0.191 9 | NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding |
| R5 | POPexmRNANahr.Psal → GFP.mRNA | 0.11 | Transcription rate of GFP in molecules/sec (for GFP size 720bp10, transcription rate in E.coli 80bp/sec8) |
| R6 | GFP.mRNA → GFP | 0.084 | Translation rate of GFP in molecules/sec (for GFP size 240aa10, translation rate in E.coli 20aa/sec8) |
| R9 | GFP.mRNA → 0 | 0.03 | Degradation rate of GFP mRNA product11 must be taken into account due to suboptimal conditions |
Results
To follow
References
1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect. Biology Letters 10.1098
2. Andrady AL (2011) Microplastics in the marine environment. Marine Pollution Bulletin 62: 1596-1605 Shala AA, Restrepo S, Gonzalez Barrios AF (2011) A network model for biofilm development in Escherichia coli K-12. Theoretical Biology and Medical Modelling 8: 34 doi:10.1186/1742-4682-8-34
3. To follow
4. To follow
5. Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR (2011) CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 81: 486-499
6. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324
Walsh DM, Lomakin A, Benedek GB, Condron MM, Teplow DB (1997) Amyloid b-Protein Fibrillogenesis: Detection of a protofibrillar intermediate. The Journal of Biological Chemistry 272: 22364–22372
 "
"