Team:Goettingen/Test
From 2012.igem.org
| (8 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{GoettingenHeader}} | ||
| + | |||
<html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" dir="ltr" lang="en"> | <html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" dir="ltr" lang="en"> | ||
<head> | <head> | ||
<meta http-equiv="Content-Type" content="text/html; charset=UTF-8"> | <meta http-equiv="Content-Type" content="text/html; charset=UTF-8"> | ||
<meta name="keywords" content="Team:Goettingen,Team:Goettingen,Team:Goettingen/Homing coli,Team:Goettingen/Human Practice/Flash coli,Team:Goettingen/Press,Team:Goettingen/Project/General information"> | <meta name="keywords" content="Team:Goettingen,Team:Goettingen,Team:Goettingen/Homing coli,Team:Goettingen/Human Practice/Flash coli,Team:Goettingen/Press,Team:Goettingen/Project/General information"> | ||
| - | + | <link rel="shortcut icon" href="https://2012.igem.org/favicon.ico"> | |
| - | + | <link rel="search" type="application/opensearchdescription+xml" href="https://2012.igem.org/wiki/opensearch_desc.php" title="2012.igem.org (English)"> | |
| - | + | <link title="Creative Commons" type="application/rdf+xml" href="https://2012.igem.org/wiki/index.php?title=Team:Goettingen&action=creativecommons" rel="meta"> | |
| - | + | <link rel="alternate" type="application/rss+xml" title="2012.igem.org RSS Feed" href="https://2012.igem.org/wiki/index.php?title=Special:Recentchanges&feed=rss"> | |
| - | + | <link rel="alternate" type="application/atom+xml" title="2012.igem.org Atom Feed" href="https://2012.igem.org/wiki/index.php?title=Special:Recentchanges&feed=atom"> | |
</head> | </head> | ||
<body class="mediawiki ns-0 ltr page-Team_Goettingen"> | <body class="mediawiki ns-0 ltr page-Team_Goettingen"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<table> | <table> | ||
| - | |||
<tbody><tr valign="top" align="left"> | <tbody><tr valign="top" align="left"> | ||
| Line 423: | Line 22: | ||
<br> | <br> | ||
| - | <table id="toc" class="toc"><tbody><tr><td><div id="toctitle"><h2>Contents</h2> <span class="toctoggle"> | + | <table id="toc" class="toc"><tbody><tr><td><div id="toctitle"><h2>Contents</h2> <span class="toctoggle"><a href="javascript:toggleToc()" class="internal" id="togglelink"></a></span></div> |
<ul> | <ul> | ||
<li class="toclevel-1"><a href="#Welcome"><span class="tocnumber">1</span> <span class="toctext">Welcome to iGEM Göttingen</span></a></li> | <li class="toclevel-1"><a href="#Welcome"><span class="tocnumber">1</span> <span class="toctext">Welcome to iGEM Göttingen</span></a></li> | ||
| Line 431: | Line 30: | ||
</td></tr></tbody></table> | </td></tr></tbody></table> | ||
<br> | <br> | ||
| - | |||
| Line 437: | Line 35: | ||
<p align="justify" style="line-height:1.6em"> | <p align="justify" style="line-height:1.6em"> | ||
| - | + | In November 2011, most of us heard about iGEM competition for the first time. | |
| - | of | + | Erik, inspired by his former colleagues´ enthusiastic reports, was looking for like-minded people at his new university that |
| - | + | were willing to organize a whole project in the field of Synthetic Biology. He described the idea of the contest to anybody willing to listen. It did not take | |
| - | + | long to find five other interested students. Thus, the so called organization task force of the first iGEM team of the University of Göttingen was formed. | |
| - | + | <br><br> | |
| - | + | Luckily, the iGEM competition was already well known among the University´s professors and the team found his first supporters: Prof. Heinz | |
| - | + | Neumann, head of a work group specializing in Synthetic Biology, and Prof. Jörg Stülke from the department of General and Applied Microbiology. | |
| - | + | The next task was to find a project that was interesting as well as realizable. After a lot of back and forth due to extensive discussions, Prof. | |
| - | + | Neumann presented the perfect project: "Homing Coli"! | |
| - | + | <br><br> | |
| - | + | After many drawbacks, like finding a suitable laboratory to realize the project, the search for additional team members began. The new | |
| - | <br> | + | fellows were quickly found, which was the story starting point of the first iGEM team of the University of Göttingen… Below you can see our timeline.<br><br> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <a href="https://static.igem.org/mediawiki/2012/0/0d/Timeline.png"><img width="650px" src="https://static.igem.org/mediawiki/2012/0/0d/Timeline.png"></a><br> | ||
| + | The timeline of the iGEM team Göttingen 2012. Click on the picture for full size. | ||
| + | <br><br> | ||
| + | If you are interested in the <a href="https://2012.igem.org/Team:Goettingen/iGEM">iGEM competition</a>, the BioBrick concept, and – most importantly – <a href="https://2012.igem.org/Team:Goettingen/Project">our project</a>, take a look around. Here, you will find everything | ||
| + | you need to know about "Homing Coli" and you will have the opportunity to gather knowledge about chemotaxis in general! | ||
| + | <br> | ||
| + | Visit our <a href="https://2012.igem.org/Team:Goettingen/Team">team site</a> to learn more about us and our supporters and advisors! | ||
| + | And, last but not least, do not forget to check out our mascot <a href="https://2012.igem.org/Team:Goettingen/Human_Practice/Flash_coli">"Flash Coli"</a>! He always has some interesting stories to tell and needs YOUR help to fight the horrible phages! | ||
| + | <br><br> | ||
| + | Feel free to browse this page!<br><br> | ||
</p> | </p> | ||
| - | |||
<br> | <br> | ||
| + | |||
| + | |||
| + | |||
| Line 485: | Line 85: | ||
beneficial fast phenotype can be combined with the ability of this bacterium to sense | beneficial fast phenotype can be combined with the ability of this bacterium to sense | ||
specific compounds in their environments. Chemoreceptors enable it to move towards | specific compounds in their environments. Chemoreceptors enable it to move towards | ||
| - | or along gradients of such substances. We will use PCR-based site directed mutagenesis on the <i> E. coli</i> | + | or along gradients of such substances. We will use PCR-based site directed mutagenesis on the <i> E. coli</i> Tar receptor. The combination of speed and chemotaxis allows |
us to identify <i>E. coli</i> strains, which can sense interesting compounds. Thereby, an | us to identify <i>E. coli</i> strains, which can sense interesting compounds. Thereby, an | ||
easy method for the detection of pollutants, toxins or even tumors could be provided.<br><br> | easy method for the detection of pollutants, toxins or even tumors could be provided.<br><br> | ||
| Line 491: | Line 91: | ||
<table cellpadding="3"><tr><td> | <table cellpadding="3"><tr><td> | ||
<img width="400 px" src="http://www.patrickreinke.de/igem/homingcoli.jpg"></td> | <img width="400 px" src="http://www.patrickreinke.de/igem/homingcoli.jpg"></td> | ||
| - | <td valign="top"; cellpadding="30"><font size="- | + | <td valign="top"; cellpadding="30"><font size="-2"> <a href="http://jb.asm.org/content/186/22/7529.full"> <p align="justify">Clive S. Barker <i> et al. </i> (2004). Increased Motility of <i> Escherichia coli </i> by Insertion Sequence Element |
Integration into the Regulatory Region of the flhD Operon. Journal of Bacteriology, Vol. 186: 7529-7537.</a> </font><br> | Integration into the Regulatory Region of the flhD Operon. Journal of Bacteriology, Vol. 186: 7529-7537.</a> </font><br> | ||
</td></tr> | </td></tr> | ||
| Line 498: | Line 98: | ||
<br> | <br> | ||
| + | <font size="-1"> | ||
<h2><b><a name="Synthetic_Biology"></a>Synthetic Biology</b></h2> | <h2><b><a name="Synthetic_Biology"></a>Synthetic Biology</b></h2> | ||
| Line 519: | Line 120: | ||
<img src="http://www.patrickreinke.de/igem/synbio.jpg"> | <img src="http://www.patrickreinke.de/igem/synbio.jpg"> | ||
<br> | <br> | ||
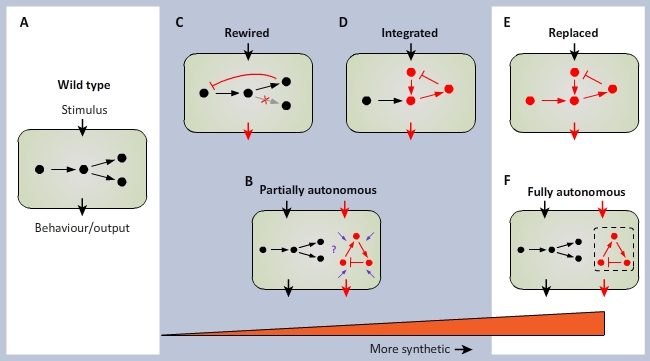
| - | <font size="- | + | <font size="-2">After Nandagopaland and Elowitz (2011). A continuum of synthetic biology. Wild-type cells (<b>A</b>) |
| - | + | can be subject to two basic types of synthetic manipulation. (<b>B</b>) Autonomous synthetic circuits, consisting of | |
| - | + | ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (red arrows) | |
| - | + | seperate from the endogenous circuitry (black). However, unknown interactions with the host cell may affect their function | |
| + | (purple arrows) (<b>C</b>)An alternative is to rewire (red lines) the endogenous circuits themselves to have new connectivity. | ||
| + | (<b>D</b>) Extending this line of synthetic manipulation, synthetic circuits could be integrated into appropriately rewired | ||
| + | endogenous circuitry to act as sensors and to implent additional functionality. Ultimate goals of this program are to be | ||
| + | able to design and construct (<b>E</b>) synthetic circuits that can functionality replace endogenous circuits or (<b>F</b>) | ||
| + | fully autonomous circuits that operate independently of the cellular mileu. | ||
| + | <a href="http://www.its.caltech.edu/~haylab/research/SBReview2011.pdf">Nagarajan Nandagopal and Michael B. Elowitz. (2011). Synthetic Biology: Integrated | ||
Gene Circuits. SCIENCE, Vol. 333: 1244-1248.</a> </font><br> | Gene Circuits. SCIENCE, Vol. 333: 1244-1248.</a> </font><br> | ||
<br> | <br> | ||
| + | <font size="-1"> | ||
Another very important and necessary feature of biological parts is orthogonality, | Another very important and necessary feature of biological parts is orthogonality, | ||
which means in this context that independent devices can be combined unrestrictedly. | which means in this context that independent devices can be combined unrestrictedly. | ||
| Line 581: | Line 189: | ||
<hr><h2> <span><b>Flash Coli</b></span></h2> | <hr><h2> <span><b>Flash Coli</b></span></h2> | ||
<!-- News cap end --> | <!-- News cap end --> | ||
| - | <embed src="http://www.patrickreinke.de/igem/motionflashcoli.swf" type="application/x-shockwave-flash" pluginspage="http://www.macromedia.com/go/getflashplayer" wmode="transparent" height="180" width="240"> | + | <embed src="http://www.patrickreinke.de/igem/motionflashcoli.swf" type="application/x-shockwave-flash" |
| + | pluginspage="http://www.macromedia.com/go/getflashplayer" wmode="transparent" height="180" width="240"> | ||
<font face="Verdana" size="-1">To explore our project with Flash Coli click | <font face="Verdana" size="-1">To explore our project with Flash Coli click | ||
<a href="https://2012.igem.org/Team:Goettingen/Human_Practice/Flash_coli">here</a>. | <a href="https://2012.igem.org/Team:Goettingen/Human_Practice/Flash_coli">here</a>. | ||
| Line 601: | Line 210: | ||
<br> | <br> | ||
<!-- News 17 --> | <!-- News 17 --> | ||
| - | <p align="left"><i>07 | + | <p align="left"><i>07 September 2012,</i> <b>We organized the "Legoland for Scientists" symposium at the University of Göttingen. |
| + | You can read everything about it <a href="https://2012.igem.org/Team:Goettingen/Human_Practice/Panel_Discussion">here.</a></b> | ||
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#17">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#17">To the news</a><br> | ||
| Line 607: | Line 217: | ||
<!-- News 16 --> | <!-- News 16 --> | ||
| - | <p align="left"><i>30 August 2012,</i> <b>The first version of | + | <p align="left"><i>30 August 2012,</i> <b>The first version of "Flash Coli - the game" is now online. Click |
| + | <a href="https://2012.igem.org/Team:Goettingen/Human_Practice/Flash_coli">here.</a></b> | ||
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#15">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#15">To the news</a><br> | ||
| Line 613: | Line 224: | ||
<!-- News 15 --> | <!-- News 15 --> | ||
| - | <p align="left"><i>25 August 2012,</i> <b>Today is the " | + | <p align="left"><i>25 August 2012,</i> <b>Today is the "Synthetic Biology" day in Germany!</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#15">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#15">To the news</a><br> | ||
| Line 619: | Line 230: | ||
<!-- News 14 --> | <!-- News 14 --> | ||
| - | <p align="left"><i>15 August 2012,</i> <b>In our weekly seminar Anna presented an example for | + | <p align="left"><i>15 August 2012,</i> <b>In our weekly seminar Anna presented an example for Synthetic Biology.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#14">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#14">To the news</a><br> | ||
| Line 627: | Line 238: | ||
<!-- News 13 --> | <!-- News 13 --> | ||
| - | <p align="left"><i>08 August 2012,</i> <b> | + | <p align="left"><i>08 August 2012,</i> <b> Survey: Ready to be asked? </b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#13">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#13">To the news</a><br> | ||
<!-- News 13 end--><br><hr> | <!-- News 13 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 12.75 --> | ||
| + | <p align="left"><i>01 August 2012,</i> <b>Ali and Sandra conducted their presentations in our weekly seminar.</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#12.75">To the news</a><br> | ||
| + | <!-- News 12.75 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 12.5 --> | ||
| + | <p align="left"><i>19 August 2012,</i> <b>Human practice task force established!</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#12.5">To the news</a><br> | ||
| + | <!-- News 12.5 end--><br><hr> | ||
<!-- News 12 --> | <!-- News 12 --> | ||
| - | <p align="left"><i>12 July 2012,</i> <b>In today’s weekly meeting Christian told us about the European teacher meet-up in Paris</b> | + | <p align="left"><i>12 July 2012,</i> <b>In today’s weekly meeting Christian told us about the European teacher meet-up in Paris.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#12">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#12">To the news</a><br> | ||
<!-- News 12 end--><br><hr> | <!-- News 12 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 11.5 --> | ||
| + | <p align="left"><i>04 July 2012,</i> <b>Let there be light!</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#11.5">To the news</a><br> | ||
| + | <!-- News 11.5 end--><br><hr> | ||
| + | |||
<!-- News 11 --> | <!-- News 11 --> | ||
| - | <p align="left"><i>28 June 2012,</i> <b>Anna, Corinna and Erik participated in the third annual congress for strategy processes | + | <p align="left"><i>28 June 2012,</i> <b>Anna, Corinna and Erik participated in the third annual congress for strategy processes "Biotechnology 2020+".</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#11">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#11">To the news</a><br> | ||
<!-- News 11 end--><br><hr> | <!-- News 11 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 10.5 --> | ||
| + | <p align="left"><i>26 June 2012,</i> <b><i>E. coli</i> strain spurred!</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#10.5">To the news</a><br> | ||
| + | <!-- News 10.5 end--><br><hr> | ||
<!-- News 10 --> | <!-- News 10 --> | ||
| - | <p align="left"><i>21 June 2012,</i> <b>Our first poster is designed</b> | + | <p align="left"><i>21 June 2012,</i> <b>Our first poster is designed!</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#10">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#10">To the news</a><br> | ||
<!-- News 10 end--><br><hr> | <!-- News 10 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 9.5 --> | ||
| + | <p align="left"><i>13 June 2012,</i> <b>Task: Find a human practice project!</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#9.5">To the news</a><br> | ||
| + | <!-- News 9.5 end--><br><hr> | ||
<!-- News 9 --> | <!-- News 9 --> | ||
| - | <p align="left"><i>06 June 2012,</i> <b>Thomas Maschner from LMU Munich came to visit</b> | + | <p align="left"><i>06 June 2012,</i> <b>Thomas Maschner, advisor from the iGEM team of the LMU Munich, came to visit us.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#9">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#9">To the news</a><br> | ||
<!-- News 9 end--><br><hr> | <!-- News 9 end--><br><hr> | ||
| + | |||
| + | |||
| + | |||
| + | <!-- News 8.5 --> | ||
| + | <p align="left"><i>01 June 2012,</i> <b>Discussion of the first lab results</b> | ||
| + | </p> | ||
| + | <a href="https://2012.igem.org/Team:Goettingen/News#8.5">To the news</a><br> | ||
| + | <!-- News 8.5 end--><br><hr> | ||
<!-- News 8 --> | <!-- News 8 --> | ||
| - | <p align="left"><i>15 May 2012,</i> <b>The first picture of our complete | + | <p align="left"><i>15 May 2012,</i> <b>The first picture of our complete team.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#8">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#8">To the news</a><br> | ||
| Line 675: | Line 335: | ||
<!-- News 7 --> | <!-- News 7 --> | ||
| - | <p align="left"><i>11 May 2012,</i> <b>iGEM Göttingen is in the local | + | <p align="left"><i>11 May 2012,</i> <b>iGEM Göttingen is in the local newspaper.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#7">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#7">To the news</a><br> | ||
| Line 683: | Line 343: | ||
<!-- News 6 --> <br> | <!-- News 6 --> <br> | ||
| - | <p align="left"><i>04 May 2012,</i> <b><a href="http://www.kws.com/">KWS SAAT AG</a> is now Head Sponsor</b> | + | <p align="left"><i>04 May 2012,</i> <b><a href="http://www.kws.com/">KWS SAAT AG</a> is now Head Sponsor.</b> |
</p> | </p> | ||
<p align="left"> | <p align="left"> | ||
| Line 693: | Line 353: | ||
<!-- News 5 --> | <!-- News 5 --> | ||
| - | <p align="left"><i>02 May 2012,</i> <b>Today Anna presented our iGEM | + | <p align="left"><i>02 May 2012,</i> <b>Today Anna presented our iGEM logo.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#5">To the news</a><br> | ||
| Line 701: | Line 361: | ||
<!-- News 4 --> | <!-- News 4 --> | ||
| - | <p align="left"><i>30 April 2012,</i> <b>We inspect the | + | <p align="left"><i>30 April 2012,</i> <b>We inspect the laboratory at the Max Planck campus.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#4">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#4">To the news</a><br> | ||
| Line 709: | Line 369: | ||
<!-- News 3 --> | <!-- News 3 --> | ||
| - | <p align="left"><i>26 April 2012,</i> <b>Weekly meeting: PCR-Lecture from Heinz</b> | + | <p align="left"><i>26 April 2012,</i> <b>Weekly meeting: PCR-Lecture from Heinz.</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#3">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#3">To the news</a><br> | ||
| Line 717: | Line 377: | ||
<!-- News 2 --> | <!-- News 2 --> | ||
| - | <p align="left"><i>19 April 2012,</i> <b>The iGEM | + | <p align="left"><i>19 April 2012,</i> <b>The iGEM team Göttingen 2012 is now complete!</b> |
</p> | </p> | ||
<a href="https://2012.igem.org/Team:Goettingen/News#2">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#2">To the news</a><br> | ||
| Line 725: | Line 385: | ||
<!-- News 1 --> <br> | <!-- News 1 --> <br> | ||
| - | </p><p><i>04 April 2012,</i> <b> <a href="http://www.sartorius.com/">Sartorius Stedim Biotech GmbH</a> is now Premium Sponsor</b> | + | </p><p><i>04 April 2012,</i> <b> <a href="http://www.sartorius.com/">Sartorius Stedim Biotech GmbH</a> is now Premium Sponsor.</b> |
</p> | </p> | ||
<p> | <p> | ||
| Line 734: | Line 394: | ||
<br> | <br> | ||
</font> | </font> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</font></td></tr></tbody> | </font></td></tr></tbody> | ||
| - | |||
</table></div> | </table></div> | ||
| - | |||
</td></tr> | </td></tr> | ||
| - | + | </body> | |
| - | {{ | + | </html> |
| + | {{GoettingenFooter}} | ||
Latest revision as of 14:03, 20 September 2012
 | |
Deutsch  / English / English  |
 |
|
Language: Welcome to iGEM Göttingen
In November 2011, most of us heard about iGEM competition for the first time.
Erik, inspired by his former colleagues´ enthusiastic reports, was looking for like-minded people at his new university that
were willing to organize a whole project in the field of Synthetic Biology. He described the idea of the contest to anybody willing to listen. It did not take
long to find five other interested students. Thus, the so called organization task force of the first iGEM team of the University of Göttingen was formed.
Homing Coli
Escherichia coli is a commonly used bacterial model organism. It has lots of beneficial
traits, e.g. a short generation time and it can be easily manipulated. Most E. coli
strains that are used in laboratories do not exhibit high motility. The crucial element for
motility is the flagellum, which is rotated by a molecular motor within the cell wall.
Consequently, these are reduced in cultivated E. coli strains. Synthetic Biology
Synthetic biology is an interdisciplinary scientific area that has recently developed.
It links various fields of science like biology, chemistry, physics, molecular genetics,
informatics and engineering. ↑ Return to top
|
Flash Coli |
| ↑ Back to top! |

|
 "
"