Team:UNAM Genomics Mexico/Modeling/OR
From 2012.igem.org
| (11 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
<br /> | <br /> | ||
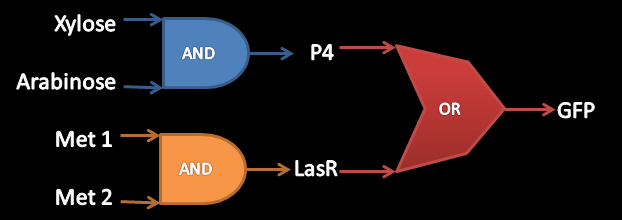
| - | Ok, this is the good one. For the final step, we had two pretty AND gates that had a special response for certain states of their inputs, but we wanted more. So, how could we make a bigger circuit? The trivial answer, obviously, was to wire everything together under a NEW gate! The new one makes an OR operation, where its inputs are the previous gates' outputs. Both outputs, P4 and LasR are reported activators for | + | Ok, this is the good one. For the final step, we had two pretty AND gates that had a special response for certain states of their inputs, but we wanted more. So, how could we make a bigger circuit? The trivial answer, obviously, was to wire everything together under a NEW gate! The new one makes an OR operation, where its inputs are the previous gates' outputs. Both outputs, P4 and LasR, are reported activators for promoters A3 and LasB respectively. In that way, our OR consists of two independent promoters that have the same reporter protein downstream: GFP. <br /> |
<br /> | <br /> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnblack" align= "center"><br />[[File:UGM_OR.png]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| - | [[File: | + | <center> |
| - | <br /> | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> |
| - | [ | + | <tr> |
| + | <td id="rightcolumn2" align= "center"><br />[[File:UGM_tv_or.png]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | |||
| + | |||
| + | |||
| + | According to both approaches, Stochastic and deterministic, the Or behaves as an OR. [https://static.igem.org/mediawiki/2012/a/a5/OR.txt Kappa OR File]<br /> | ||
<br /> | <br /> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:P41000_las0.png | 400px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:P410_las1000.png | 400px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| - | + | <center> | |
| - | [ | + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> |
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:Ep41000_las1000.png | 400px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:UNAM_GENOMICS_MEXICO_OR_3D_3D.png | 850px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | For the deterministic approach, we decided to use the Hill function designed for activators. Being additive events, the production of reporter mRNA will be the sum of the productions by each promoter, as defined by their respective affinities and intracellular concentrations. Phi and Omega are the cooperative binding rate given by the activator [2]. <br /> | ||
| + | <br /> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:UGM_O10.png]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | |||
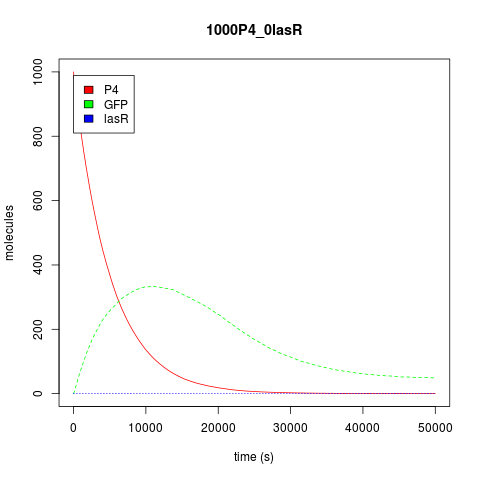
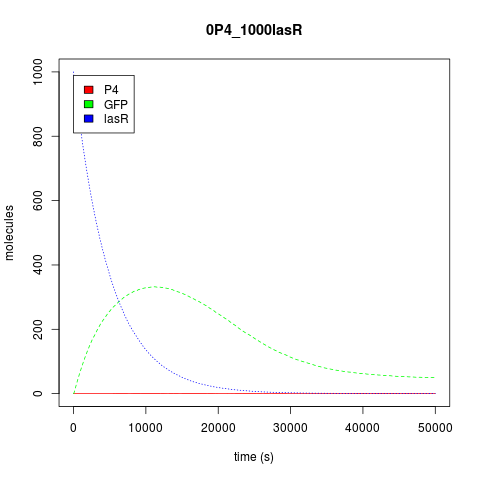
| + | Now, about the behavior of this construction predicted by our model, assuming that both transcription factors had the same affinity, both the stochastic and the deterministic approach show us that its behavior is not unlike an Or gate. As seen from the perspective of the transcription factors, activation occurs when any of them is at high concentration in the system. As the OR is activated by two different promoters, activation does seem to reach its summit when there are both activators in the system, while the genes are only basally expressed when no activator is present. <br /> | ||
| + | |||
| + | ::::::::::::::::::::::::::::::::::::::[[File:UnamgenomcisUp.png|right | 120px |link=Team:UNAM_Genomics_Mexico/Modeling/OR#OR_Results]] | ||
| + | References: <br /> | ||
| + | [1] Buchler et al. On schemes of combinatorial transcription logic print April 17, 2003, doi: 10.1073/pnas.0930314100 PNAS April 29, 2003 vol. 100 no. 9 5136-5141<br /> | ||
| + | <br /> | ||
| + | [2]Bintu L, Buchler N, Garcia H G, Gerland U, Hwa T. Transcriptional regulation by the numbers: Models. Genetics & Development. Opinion. 2005. 15(2):116-124.DOI:10.1016<br /> | ||
| + | <br /> | ||
}} | }} | ||
Latest revision as of 04:00, 27 October 2012

OR Results
OR gate
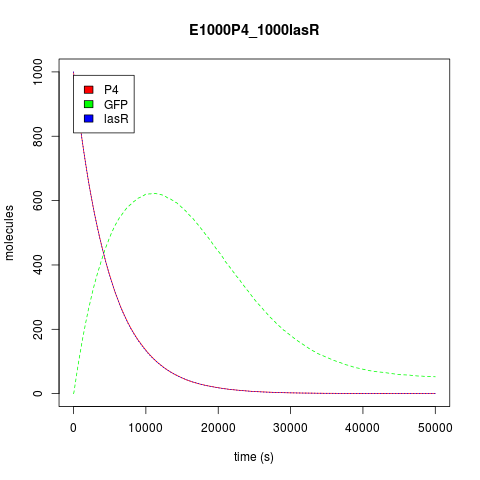
Ok, this is the good one. For the final step, we had two pretty AND gates that had a special response for certain states of their inputs, but we wanted more. So, how could we make a bigger circuit? The trivial answer, obviously, was to wire everything together under a NEW gate! The new one makes an OR operation, where its inputs are the previous gates' outputs. Both outputs, P4 and LasR, are reported activators for promoters A3 and LasB respectively. In that way, our OR consists of two independent promoters that have the same reporter protein downstream: GFP.
|
|
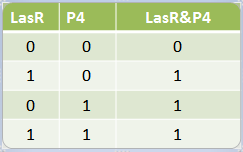
According to both approaches, Stochastic and deterministic, the Or behaves as an OR. Kappa OR File
|
|
|
|
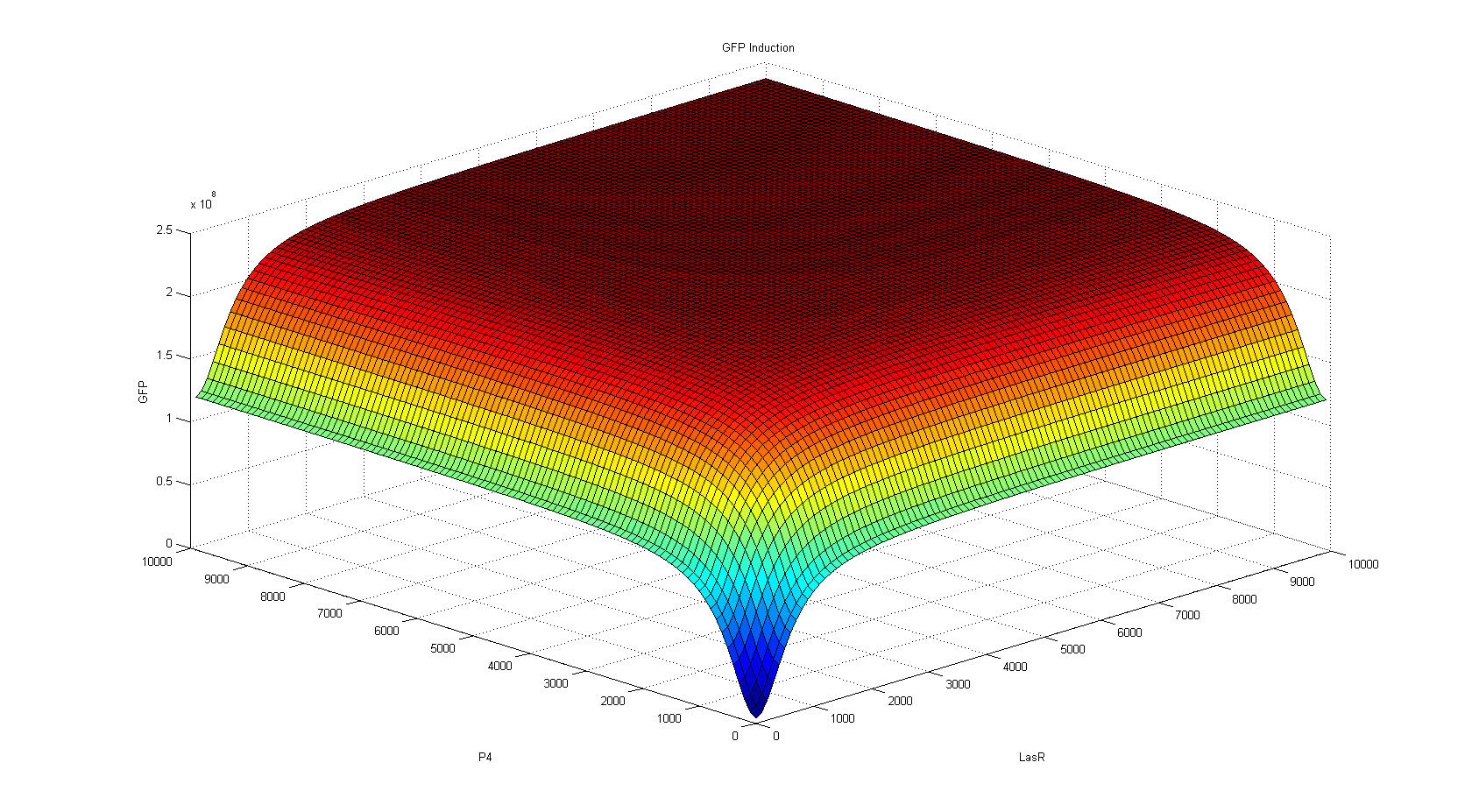
For the deterministic approach, we decided to use the Hill function designed for activators. Being additive events, the production of reporter mRNA will be the sum of the productions by each promoter, as defined by their respective affinities and intracellular concentrations. Phi and Omega are the cooperative binding rate given by the activator [2].

|
Now, about the behavior of this construction predicted by our model, assuming that both transcription factors had the same affinity, both the stochastic and the deterministic approach show us that its behavior is not unlike an Or gate. As seen from the perspective of the transcription factors, activation occurs when any of them is at high concentration in the system. As the OR is activated by two different promoters, activation does seem to reach its summit when there are both activators in the system, while the genes are only basally expressed when no activator is present.
References:
[1] Buchler et al. On schemes of combinatorial transcription logic print April 17, 2003, doi: 10.1073/pnas.0930314100 PNAS April 29, 2003 vol. 100 no. 9 5136-5141
[2]Bintu L, Buchler N, Garcia H G, Gerland U, Hwa T. Transcriptional regulation by the numbers: Models. Genetics & Development. Opinion. 2005. 15(2):116-124.DOI:10.1016
 "
"







