Team:TU-Delft/Modeling/SingleCellModel
From 2012.igem.org

The single-cell pathway model of our system is composed of 4 modules, which are responsible for routing the signal from the receptor to the GFP sythesis. The mathematical models were developed on a scheme favoring the temporal order of processes, based on the current understanding of the pheromone signalling pathway from [1][2][3] and also on the feedback received from the experimentalists on the expected behaviour of the pathway, taking into consideration the aspects relevant to our project.
Contents |
Assumptions
In all of the models developed certain assumptions were made due to the lack of data available on the new modified pathway and also due to the time constraints which limit the range of experiments that can be carried out. The assumptions made are listed below.
- The rate constants and initial concentrations obtained from the literature are assumed to be the same for the new modified pathway.
- The behaviour of the Ligand is assumed to be similar to the behaviour of the alpha pheromone.
- The tuberculosis receptor is assumed to behave similar to the alpha-pheromone receptor.
Model of the G - Protein cycle
The concentration of free G-beta-gamma dimer is crucial for the activation of the MAP kinase cascade which in turn activates the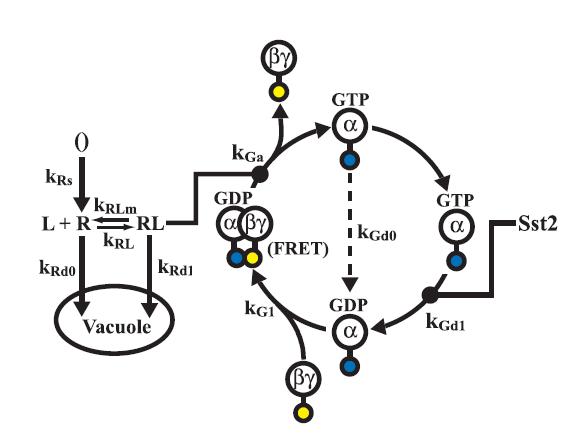
It can be seen that as the concentration of G-alpha-GDP increases the concentration of free G-beta-gamma dimer reduces, which motivates the use of a promoter which is weak or medium strength compared to a strong promoter.
Model of the Yeast Pheromone Response
The model of the yeast pheromone response was based on the model by Kofahl and Klipp[2]. This was used to test the effects of cell cycle arrest knockout on the transcription factor Ste12 which is crucial for the synthesis of GFP.
It can be seen from the plots that for an input concentration of 100nm, the knockout performed has no major influence on the concentration of the transcription factor.
Model of the Snifformyces(Modified Yeast) Pathway
The model of the G-protein cycle lacked the description of MAP kinase cascade and the gene expression module.The model of the yeast pheromone response had a detailed description of the pathway but had two drawbacks, it did not incorporate the gene expression module and more importantly was too complex in terms of the number of parameters that were present in the model. Due to the large number of parameters, fitting the limited data available from the experiments to the highly complex pathway model in [2] we felt would not have been feasible.
The above drawbacks motivated us to build a model, which would capture the crucial components of the pathway with a reduced degree of complexity i.e with lesser parameters to be estimated. Combining the knowledge from [1] and [2] and also using [3], We built a new model of the pathway. The schematic of which is in
The rates in this model were obtained by fitting the model to the data in [4]. It can be seen from the simulations that the model is capable of reproducing the Fus3 dynamics fairly accurately.
References

 "
"
