Team:TU-Delft/Modeling/SingleCellModel
From 2012.igem.org
(→Parameter Estimation) |
(→Stochastic Model) |
||
| Line 86: | Line 86: | ||
<div id="contentbox" style="text-align:justify;"> | <div id="contentbox" style="text-align:justify;"> | ||
= Stochastic Model = | = Stochastic Model = | ||
| - | In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population[[Team:TU-Delft/Modeling/SingleCellModel#Ref7|[7]]]. In order to investigate the effects of this stochasticity, we decided to build a stochastic model of the pathway. a Hybrid ODE-SDE(stochastic differential equation) model was constructed. Data from [[Team:TU-Delft/Modeling/SingleCellModel#Ref7|[5]]] suggests that the Pheromone signalling is robust against cell to cell variations, Motivated by this, we assume the Pheromone signalling to be deterministic rather than stochastic. As a result of the gene expression being noisy, we build a hybrid stochastic model consisting of deterministic semantics uptil the activation of the transcription factor Ste12 and treat it as a time varying parameter modulating the reaction based gene expression module interpreted with stochastic semantics using the stochastic differential equation approach.The schematic of which is in figure --. | + | In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population[[Team:TU-Delft/Modeling/SingleCellModel#Ref7|[7]]]. In order to investigate the effects of this stochasticity, we decided to build a stochastic model of the pathway. a '''Hybrid ODE-SDE(stochastic differential equation)''' model was constructed. Data from [[Team:TU-Delft/Modeling/SingleCellModel#Ref7|[5]]] suggests that the Pheromone signalling is robust against cell to cell variations, Motivated by this, we assume the Pheromone signalling to be deterministic rather than stochastic. As a result of the gene expression being noisy, we build a hybrid stochastic model consisting of deterministic semantics uptil the activation of the transcription factor Ste12 and treat it as a time varying parameter modulating the reaction based gene expression module interpreted with stochastic semantics using the stochastic differential equation approach.The schematic of which is in figure --. |
</div> | </div> | ||
Revision as of 21:28, 25 September 2012

The single-cell pathway model of our system is composed of 4 modules, which are responsible for routing the signal from the receptor to the GFP sythesis. The mathematical models were developed on a scheme favoring the temporal order of processes, based on the current understanding of the pheromone signalling pathway from [1][2][3] and also on the feedback received from the experimentalists on the expected behaviour of the pathway, taking into consideration the aspects relevant to our project.
Contents |
Assumptions
In all of the models developed certain assumptions were made due to the lack of data available on the new modified pathway and also due to the time constraints which limit the range of experiments that can be carried out. The assumptions made are listed below.
- The rate constants and initial concentrations obtained from the literature are assumed to be the same for the new modified pathway.
- The behaviour of the Ligand is assumed to be similar to the behaviour of the alpha pheromone.
- The receptor is assumed to behave similar to the alpha-pheromone receptor.
Model of the G - Protein cycle
The concentration of free G-beta-gamma dimer is crucial for the activation of the MAP kinase cascade which in turn activates the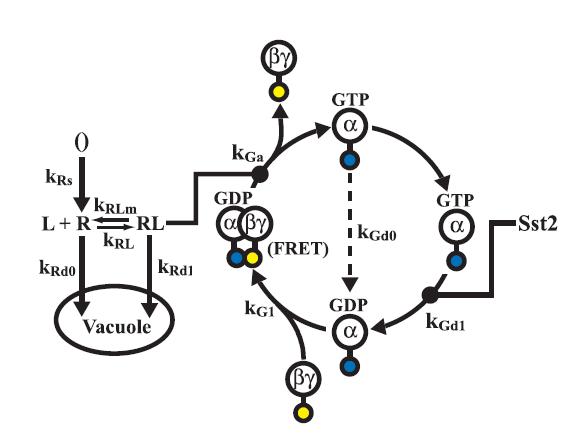
It can be seen that as the concentration of G-alpha-GDP increases the concentration of free G-beta-gamma dimer reduces, which motivates the use of a promoter which is weak or medium strength compared to a strong promoter.
Model of the Yeast Pheromone Response
The model of the yeast pheromone response was based on the model by Kofahl and Klipp[2]. This was used to test the effects of Far1 gene knockout on the transcription factor Ste12 which is crucial for the synthesis of GFP.
It can be seen from the plots that, the knockout has no major influence on the concentration of the transcription factor. This was a crucial observation which helped us go proceed with the knockout as it did not affect the concentration of the transcription factor. The rates used in the model can be found here.
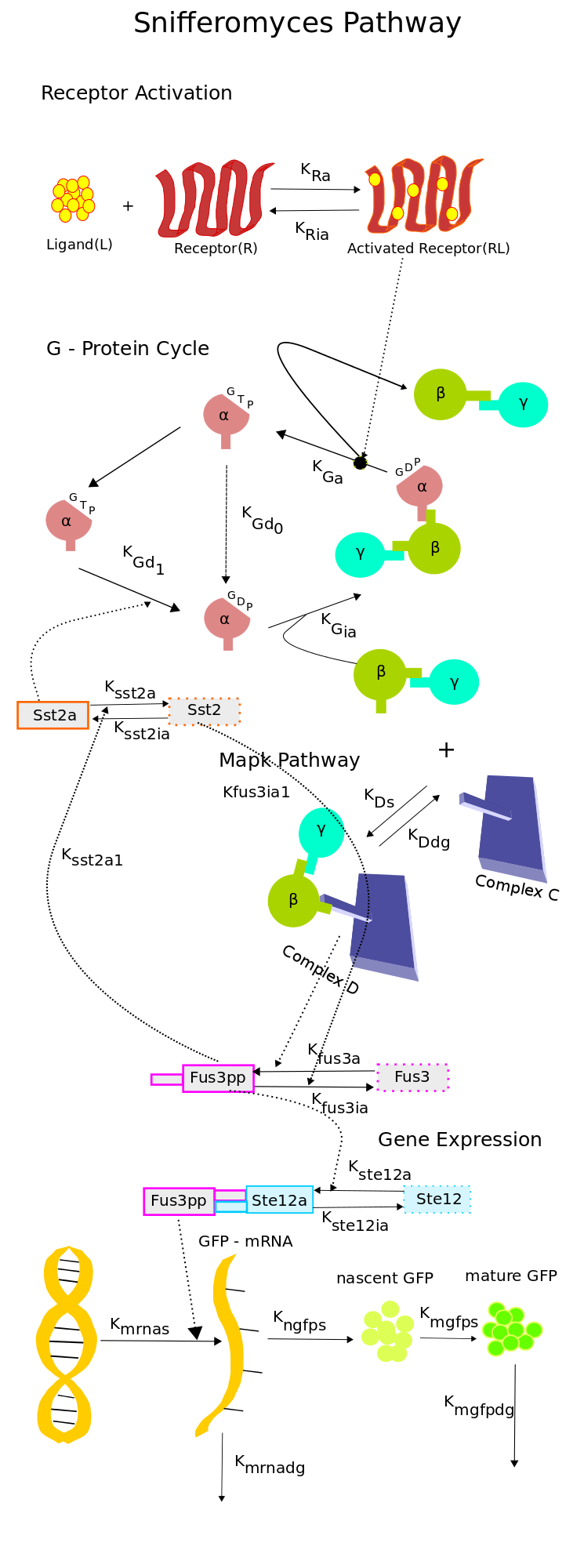
Reduced Model of the Snifformyces (Modified Yeast) Pathway
The model of the G-protein cycle lacked the description of MAP kinase cascade and the gene expression module.The model of the yeast pheromone response on the other hand had a detailed description of the pathway but had two drawbacks, it did not incorporate the gene expression module and more importantly was too complex in terms of the number of parameters that were present in the model. Due to the large number of parameters, fitting the limited data available from the experiments to the highly complex pathway model in [2] we felt would not have been feasible.The above drawbacks motivated us to build a reduced order model, which would capture the crucial components of the pathway with a reduced degree of complexity i.e with lesser parameters to be estimated. For this reason we built a new reduced ordel model of the pathway which had 18 species and 21 rate constant, which when compared to the 37 species and 47 rate constants of the model in [2] was a significant reduction.
To construct the model a mechanistic approach was taken. Combining the knowledge from [1] and [2] and also using [3] and [4], we constructed the new model shown in the schematic.
The key features of the model are as follows,
- The Receptor activation & the G - Protein cycle was retained from [1]. The ligand and the receptor degradation were neglected owing to the observation that, the receptors were placed under a constitutive promoter and ligand would not be digested by yeast.
- The simplified MAP kinase cascade was adopted from [3]. The MAPK pathway consists of a complex C to which the free G-beta-gamma dimer binds resulting in the activation of Fus3. The simplified structure captures the dynamics of the activated Fus3 in an efficient manner as can be seen in [3]. This simplification helped a great deal in eliminating the components of the pathway which were not crucial to our experiments. Since the only key components that were under our control were the ligand concentration, the promoter of the receptor and the Fus1-GFP promoter, we decided to retain the G-protein cycle in it's entirety and reduce the MAP kinase pathways to simple two stage reaction.
- The activation of Ste12 was adopted from [2].
- For the gene expression module, a reaction based model was used which was based on [4]. In this model, the mRNA synthesis is proportional to the concentration of activated Ste12 and the protein synthesis is proportional to the mRNA concentration. The protein synthesis is represented in two stages, the synthesis of nascent GFP which is proportional to the mRNA concentration and the synthesis of mature GFP which is proportional to the concentration of nascent GFP. The mRNA and mature GFP degradations are considered proportional to their concentrations.
For the initial analysis, the rates of this model uptil Fus3 activation were obtained from [3] in which the model was fit to the data from [5]. The Ste12 activation and deactivation rates were taken from [2]. The rates for the gene expression module were obtained from [4]. The initial model was first interpreted using deterministic semantics. The rates for which can be found - - - .
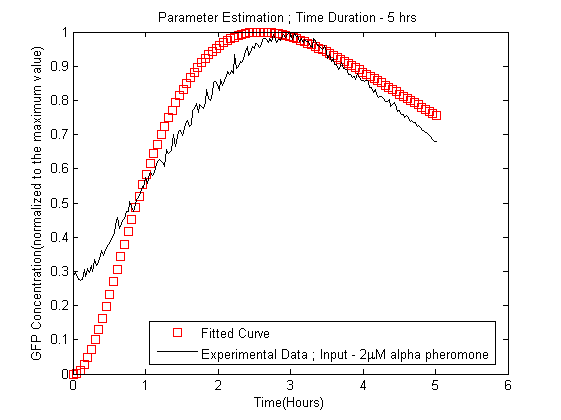
Parameter Estimation
Before the model could be used for predictions, we needed realistic parameters that suited our model, using the results from the sensitivity analysis, parameter estimation was performed to find the parameters of the model in order to fit the simulated data curves to the experimental data. The data fitting was done by using the nonlinear regression function nlinfit in the Matlab statistical toolbox[6].
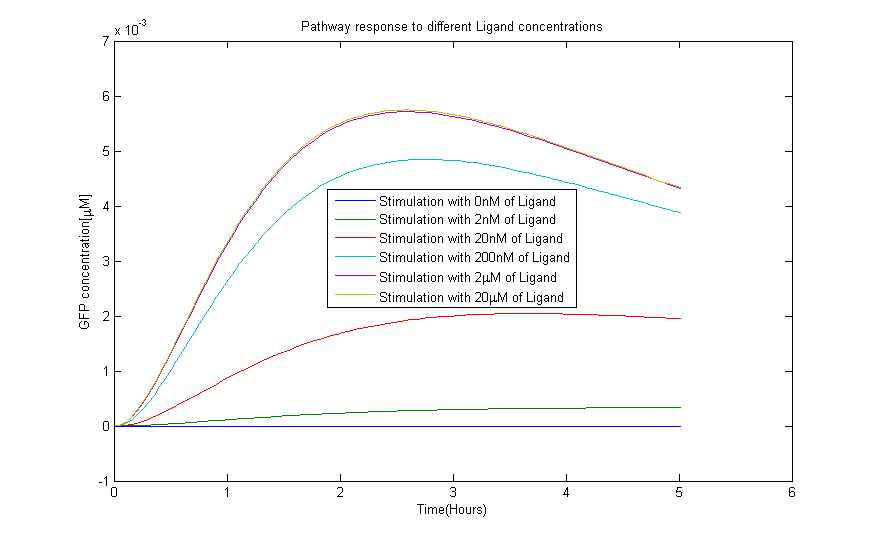
Model Predictions
The model with the new estimated parameters was then used to make predictions on the behaviour of the pathway for different concentrations of the Ligand. The model was simulated for a range of input concentrations using the ode15s variable order solver from Matlab. The results of the simulation are in Figure 7.The GFP concentration increases linearly with the induced Ligand uptil a concentration of 2 micro Molar above which the output concentration saturates. These results could also be seen from the experiments. -- Place link
Stochastic Model
In Saccharomyces cerevisiae, stochasticity (noise) arising from transcription contributes significantly to the level of heterogeneity within a eukaryotic clonal population[7]. In order to investigate the effects of this stochasticity, we decided to build a stochastic model of the pathway. a Hybrid ODE-SDE(stochastic differential equation) model was constructed. Data from [5] suggests that the Pheromone signalling is robust against cell to cell variations, Motivated by this, we assume the Pheromone signalling to be deterministic rather than stochastic. As a result of the gene expression being noisy, we build a hybrid stochastic model consisting of deterministic semantics uptil the activation of the transcription factor Ste12 and treat it as a time varying parameter modulating the reaction based gene expression module interpreted with stochastic semantics using the stochastic differential equation approach.The schematic of which is in figure --.
References
 "
"