Team:Goettingen
From 2012.igem.org
| Line 1: | Line 1: | ||
| - | {{GoettingenHeader | + | {{GoettingenHeader|deu=Team:Goettingen/main_deu|eng=Team:Goettingen}} |
| - | + | {| style="width: 900px; margin:0 0 0 0; background:none; border-spacing: 0px" | |
| - | + | | style="padding: 0pt 20px 0pt 0pt; width:630px; vertical-align:top" | | |
| - | + | == Welcome to iGEM Göttingen == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
In November 2011, most of us heard about iGEM competition for the first time. | In November 2011, most of us heard about iGEM competition for the first time. | ||
Erik, inspired by his former colleagues's enthusiastic reports, was looking for like-minded people at his new university that | Erik, inspired by his former colleagues's enthusiastic reports, was looking for like-minded people at his new university that | ||
were willing to organize a whole project in the field of Synthetic Biology. He described the idea of the contest to anybody willing to listen. It did not take | were willing to organize a whole project in the field of Synthetic Biology. He described the idea of the contest to anybody willing to listen. It did not take | ||
long to find five other interested students. Thus, the so called organization task force of the first iGEM team of the University of Göttingen was formed. | long to find five other interested students. Thus, the so called organization task force of the first iGEM team of the University of Göttingen was formed. | ||
| - | + | ||
Luckily, the iGEM competition was already well known among the University´s professors and the team found his first supporters: Prof. Heinz | Luckily, the iGEM competition was already well known among the University´s professors and the team found his first supporters: Prof. Heinz | ||
Neumann, head of a work group specializing in Synthetic Biology, and Prof. Jörg Stülke from the department of General and Applied Microbiology. | Neumann, head of a work group specializing in Synthetic Biology, and Prof. Jörg Stülke from the department of General and Applied Microbiology. | ||
The next task was to find a project that was interesting as well as realizable. After a lot of back and forth due to extensive discussions, Prof. | The next task was to find a project that was interesting as well as realizable. After a lot of back and forth due to extensive discussions, Prof. | ||
Neumann presented the perfect project: "Homing Coli"! | Neumann presented the perfect project: "Homing Coli"! | ||
| - | + | ||
After many drawbacks, like finding a suitable laboratory to realize the project, the search for additional team members began. The new | After many drawbacks, like finding a suitable laboratory to realize the project, the search for additional team members began. The new | ||
| - | fellows were quickly found, which was the story starting point of the first iGEM team of the University of Göttingen… Below you can see our timeline. | + | fellows were quickly found, which was the story starting point of the first iGEM team of the University of Göttingen… Below you can see our timeline. |
| - | + | [[File:Timeline.png|600px|thumb|center|The timeline of the iGEM team Göttingen 2012. Click on the picture for full size.]] | |
| - | The timeline of the iGEM team Göttingen 2012. Click on the picture for full size. | + | |
| - | + | If you are interested in the [[Team:Goettingen/iGEM|iGEM competition]], the BioBrick concept, and – most importantly – [[Team:Goettingen/Project|our project]], take a look around. Here, you will find everything | |
| - | + | ||
| - | If you are interested in the | + | |
you need to know about "Homing Coli" and you will have the opportunity to gather knowledge about chemotaxis in general! | you need to know about "Homing Coli" and you will have the opportunity to gather knowledge about chemotaxis in general! | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | Visit our [[Team:Goettingen/Team|team site]] to learn more about us and our supporters and advisors! | |
| + | And, last but not least, do not forget to check out our mascot [[Team:Goettingen/Human_Practice/Flash_coli|Flash Coli]]! He always has some interesting stories to tell and needs YOUR help to fight the horrible phages! | ||
| - | + | Feel free to browse this page! | |
| + | |||
| + | == Homing Coli == | ||
<i>Escherichia coli</i> is a commonly used bacterial model organism. It has lots of beneficial | <i>Escherichia coli</i> is a commonly used bacterial model organism. It has lots of beneficial | ||
traits, e.g. a short generation time and it can be easily manipulated. Most <i>E. coli</i> | traits, e.g. a short generation time and it can be easily manipulated. Most <i>E. coli</i> | ||
strains that are used in laboratories do not exhibit high motility. The crucial element for | strains that are used in laboratories do not exhibit high motility. The crucial element for | ||
motility is the flagellum, which is rotated by a molecular motor within the cell wall. | motility is the flagellum, which is rotated by a molecular motor within the cell wall. | ||
| - | Consequently, these are reduced in cultivated <i>E. coli</i> strains. | + | Consequently, these are reduced in cultivated <i>E. coli</i> strains. |
| - | + | ||
| - | Our goal is to create an <i>E. coli</i> strain with increased swimming motility on special | + | Our goal is to create an <i>E. coli</i> strain with increased swimming motility on special agar plates. Therefore, we will overexpress regulators and parts of the <i>E. coli</i> flagellum in cultivated strains in order to enhance bacterial swimming ability. The fastest <i>E. coli</i> strains will be selected and further improved. At the same time we will then be able to create an effective motility-selection method. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Now, you are probably wondering what the advantage of a fast <i> E. coli</i> might be. The | Now, you are probably wondering what the advantage of a fast <i> E. coli</i> might be. The | ||
beneficial fast phenotype can be combined with the ability of this bacterium to sense | beneficial fast phenotype can be combined with the ability of this bacterium to sense | ||
| Line 81: | Line 40: | ||
or along gradients of such substances. We will use PCR-based site directed mutagenesis on the <i> E. coli</i> Tar receptor. The combination of speed and chemotaxis allows | or along gradients of such substances. We will use PCR-based site directed mutagenesis on the <i> E. coli</i> Tar receptor. The combination of speed and chemotaxis allows | ||
us to identify <i>E. coli</i> strains, which can sense interesting compounds. Thereby, an | us to identify <i>E. coli</i> strains, which can sense interesting compounds. Thereby, an | ||
| - | easy method for the detection of pollutants, toxins or even tumors could be provided. | + | easy method for the detection of pollutants, toxins or even tumors could be provided. |
| - | + | [[File:Homingcoli.jpg|600px|thumb|center|<html><a href="http://jb.asm.org/content/186/22/7529.full">Clive S. Barker <i> et al. </i> (2004). Increased Motility of <i> Escherichia coli </i> by Insertion Sequence Element Integration into the Regulatory Region of the flhD Operon. Journal of Bacteriology, Vol. 186: 7529-7537.</a></html>]] | |
| - | + | ||
| - | + | ||
| - | Integration into the Regulatory Region of the flhD Operon. Journal of Bacteriology, Vol. 186: 7529-7537.</a> </ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | == Synthetic Biology == | |
| - | + | ||
| - | |||
Synthetic biology is an interdisciplinary scientific area that has recently developed. | Synthetic biology is an interdisciplinary scientific area that has recently developed. | ||
It links various fields of science like biology, chemistry, physics, molecular genetics, | It links various fields of science like biology, chemistry, physics, molecular genetics, | ||
| - | informatics and engineering. | + | informatics and engineering. |
| - | + | ||
| - | + | ||
Due to this combination the relation of biological design and function can be investigated | Due to this combination the relation of biological design and function can be investigated | ||
| Line 109: | Line 57: | ||
modular design that facilitates their handling. The subsequent introduction of the synthetic | modular design that facilitates their handling. The subsequent introduction of the synthetic | ||
constructs to living cells can either cause the replacement of original cellular components | constructs to living cells can either cause the replacement of original cellular components | ||
| - | or result in additional elements that act cooperatively or more or less autonomously (see fig. 1). | + | or result in additional elements that act cooperatively or more or less autonomously (see fig. 1). |
| - | + | ||
| - | + | [[File:SynbioGoe.jpg|600px|thumb|center|After Nandagopaland and Elowitz (2011). A continuum of synthetic biology. Wild-type cells (<b>A</b>) | |
| - | + | ||
| - | + | ||
can be subject to two basic types of synthetic manipulation. (<b>B</b>) Autonomous synthetic circuits, consisting of | can be subject to two basic types of synthetic manipulation. (<b>B</b>) Autonomous synthetic circuits, consisting of | ||
ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (red arrows) | ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (red arrows) | ||
| Line 122: | Line 67: | ||
endogenous circuitry to act as sensors and to implent additional functionality. Ultimate goals of this program are to be | endogenous circuitry to act as sensors and to implent additional functionality. Ultimate goals of this program are to be | ||
able to design and construct (<b>E</b>) synthetic circuits that can functionality replace endogenous circuits or (<b>F</b>) | able to design and construct (<b>E</b>) synthetic circuits that can functionality replace endogenous circuits or (<b>F</b>) | ||
| - | fully autonomous circuits that operate independently of the cellular mileu. | + | fully autonomous circuits that operate independently of the cellular mileu. <html><a href="http://www.its.caltech.edu/~haylab/research/SBReview2011.pdf">Nagarajan Nandagopal and Michael B. Elowitz. (2011). Synthetic Biology: Integrated Gene Circuits. SCIENCE, Vol. 333: 1244-1248.</a></html>]] |
| - | <a href="http://www.its.caltech.edu/~haylab/research/SBReview2011.pdf">Nagarajan Nandagopal and Michael B. Elowitz. (2011). Synthetic Biology: Integrated | + | |
| - | Gene Circuits. SCIENCE, Vol. 333: 1244-1248.</a> </ | + | Another very important and necessary feature of biological parts is orthogonality, which means in this context that independent devices can be combined unrestrictedly. |
| - | + | This principle derives from the area of engineering and aims to alter subsystems, without impeding others. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
This way cells can be modified as requested, resulting in a predictable behaviour. | This way cells can be modified as requested, resulting in a predictable behaviour. | ||
Among others, this technique can be beneficial for the effective production of | Among others, this technique can be beneficial for the effective production of | ||
certain substances, like biofuels or antibiotics. | certain substances, like biofuels or antibiotics. | ||
| - | + | <html><center> | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<table bordercolor="black" border="1 px" width="600 px"><tr><td> | <table bordercolor="black" border="1 px" width="600 px"><tr><td> | ||
<b>Important pages</b>:<br> | <b>Important pages</b>:<br> | ||
| Line 157: | Line 90: | ||
</td></tr> | </td></tr> | ||
</table> | </table> | ||
| - | </ | + | </center></html> |
| - | + | | style="width:240px; vertical-align:top" | | |
| - | + | <html> | |
| - | </ | + | |
| - | + | ||
| - | + | ||
| - | < | + | |
<h2><hr><span><b>Flash Coli</b></span></h2> | <h2><hr><span><b>Flash Coli</b></span></h2> | ||
<!-- News cap end --> | <!-- News cap end --> | ||
| Line 208: | Line 137: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#14">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#14">To the news</a><br> | ||
<!-- News 14 end--><br><hr> | <!-- News 14 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 13 --> | <!-- News 13 --> | ||
| Line 216: | Line 143: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#13">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#13">To the news</a><br> | ||
<!-- News 13 end--><br><hr> | <!-- News 13 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 12.75 --> | <!-- News 12.75 --> | ||
| Line 224: | Line 149: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#12.75">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#12.75">To the news</a><br> | ||
<!-- News 12.75 end--><br><hr> | <!-- News 12.75 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 12.5 --> | <!-- News 12.5 --> | ||
| Line 232: | Line 155: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#12.5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#12.5">To the news</a><br> | ||
<!-- News 12.5 end--><br><hr> | <!-- News 12.5 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 12 --> | <!-- News 12 --> | ||
| Line 240: | Line 161: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#12">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#12">To the news</a><br> | ||
<!-- News 12 end--><br><hr> | <!-- News 12 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 11.5 --> | <!-- News 11.5 --> | ||
| Line 248: | Line 167: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#11.5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#11.5">To the news</a><br> | ||
<!-- News 11.5 end--><br><hr> | <!-- News 11.5 end--><br><hr> | ||
| - | |||
| - | |||
| - | |||
<!-- News 11 --> | <!-- News 11 --> | ||
| Line 257: | Line 173: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#11">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#11">To the news</a><br> | ||
<!-- News 11 end--><br><hr> | <!-- News 11 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 10.5 --> | <!-- News 10.5 --> | ||
| Line 265: | Line 179: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#10.5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#10.5">To the news</a><br> | ||
<!-- News 10.5 end--><br><hr> | <!-- News 10.5 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 10 --> | <!-- News 10 --> | ||
| Line 273: | Line 185: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#10">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#10">To the news</a><br> | ||
<!-- News 10 end--><br><hr> | <!-- News 10 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 9.5 --> | <!-- News 9.5 --> | ||
| Line 281: | Line 191: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#9.5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#9.5">To the news</a><br> | ||
<!-- News 9.5 end--><br><hr> | <!-- News 9.5 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 9 --> | <!-- News 9 --> | ||
| Line 289: | Line 197: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#9">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#9">To the news</a><br> | ||
<!-- News 9 end--><br><hr> | <!-- News 9 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 8.5 --> | <!-- News 8.5 --> | ||
| Line 297: | Line 203: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#8.5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#8.5">To the news</a><br> | ||
<!-- News 8.5 end--><br><hr> | <!-- News 8.5 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 8 --> | <!-- News 8 --> | ||
| Line 305: | Line 209: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#8">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#8">To the news</a><br> | ||
<!-- News 8 end--><br><hr> | <!-- News 8 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 7 --> | <!-- News 7 --> | ||
| Line 313: | Line 215: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#7">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#7">To the news</a><br> | ||
<!-- News 7 end--><br><hr> | <!-- News 7 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 6 --> <br> | <!-- News 6 --> <br> | ||
| Line 323: | Line 223: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#6">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#6">To the news</a><br> | ||
<!-- News 6 end--><br><hr> | <!-- News 6 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 5 --> | <!-- News 5 --> | ||
| Line 331: | Line 229: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#5">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#5">To the news</a><br> | ||
<!-- News 5 end--><br><hr> | <!-- News 5 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 4 --> | <!-- News 4 --> | ||
| Line 339: | Line 235: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#4">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#4">To the news</a><br> | ||
<!-- News 4 end--><br><hr> | <!-- News 4 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 3 --> | <!-- News 3 --> | ||
| Line 347: | Line 241: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#3">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#3">To the news</a><br> | ||
<!-- News 3 end--><br><hr> | <!-- News 3 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 2 --> | <!-- News 2 --> | ||
| Line 355: | Line 247: | ||
<a href="https://2012.igem.org/Team:Goettingen/News#2">To the news</a><br> | <a href="https://2012.igem.org/Team:Goettingen/News#2">To the news</a><br> | ||
<!-- News 2 end--><br><hr> | <!-- News 2 end--><br><hr> | ||
| - | |||
| - | |||
<!-- News 1 --> <br> | <!-- News 1 --> <br> | ||
| Line 366: | Line 256: | ||
<!-- News 1 end--> | <!-- News 1 end--> | ||
</font> | </font> | ||
| - | |||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
| + | |} | ||
{{GoettingenFooter}} | {{GoettingenFooter}} | ||
Revision as of 13:28, 21 September 2012
 | |
Deutsch  / English / English  |
 |
Welcome to iGEM GöttingenIn November 2011, most of us heard about iGEM competition for the first time. Erik, inspired by his former colleagues's enthusiastic reports, was looking for like-minded people at his new university that were willing to organize a whole project in the field of Synthetic Biology. He described the idea of the contest to anybody willing to listen. It did not take long to find five other interested students. Thus, the so called organization task force of the first iGEM team of the University of Göttingen was formed. Luckily, the iGEM competition was already well known among the University´s professors and the team found his first supporters: Prof. Heinz Neumann, head of a work group specializing in Synthetic Biology, and Prof. Jörg Stülke from the department of General and Applied Microbiology. The next task was to find a project that was interesting as well as realizable. After a lot of back and forth due to extensive discussions, Prof. Neumann presented the perfect project: "Homing Coli"! After many drawbacks, like finding a suitable laboratory to realize the project, the search for additional team members began. The new fellows were quickly found, which was the story starting point of the first iGEM team of the University of Göttingen… Below you can see our timeline. If you are interested in the iGEM competition, the BioBrick concept, and – most importantly – our project, take a look around. Here, you will find everything you need to know about "Homing Coli" and you will have the opportunity to gather knowledge about chemotaxis in general! Visit our team site to learn more about us and our supporters and advisors! And, last but not least, do not forget to check out our mascot Flash Coli! He always has some interesting stories to tell and needs YOUR help to fight the horrible phages! Feel free to browse this page! Homing ColiEscherichia coli is a commonly used bacterial model organism. It has lots of beneficial traits, e.g. a short generation time and it can be easily manipulated. Most E. coli strains that are used in laboratories do not exhibit high motility. The crucial element for motility is the flagellum, which is rotated by a molecular motor within the cell wall. Consequently, these are reduced in cultivated E. coli strains. Our goal is to create an E. coli strain with increased swimming motility on special agar plates. Therefore, we will overexpress regulators and parts of the E. coli flagellum in cultivated strains in order to enhance bacterial swimming ability. The fastest E. coli strains will be selected and further improved. At the same time we will then be able to create an effective motility-selection method. Now, you are probably wondering what the advantage of a fast E. coli might be. The beneficial fast phenotype can be combined with the ability of this bacterium to sense specific compounds in their environments. Chemoreceptors enable it to move towards or along gradients of such substances. We will use PCR-based site directed mutagenesis on the E. coli Tar receptor. The combination of speed and chemotaxis allows us to identify E. coli strains, which can sense interesting compounds. Thereby, an easy method for the detection of pollutants, toxins or even tumors could be provided. Synthetic BiologySynthetic biology is an interdisciplinary scientific area that has recently developed. It links various fields of science like biology, chemistry, physics, molecular genetics, informatics and engineering. Due to this combination the relation of biological design and function can be investigated from an entirely new perspective. The previous approach was limited to the examination of a structure and its function and the attempt to explain how they correlate. Recently, the reverse strategy is applied. Biological parts are specifically designed and constructed according to a desired function. These parts are characterized by a standardized modular design that facilitates their handling. The subsequent introduction of the synthetic constructs to living cells can either cause the replacement of original cellular components or result in additional elements that act cooperatively or more or less autonomously (see fig. 1). 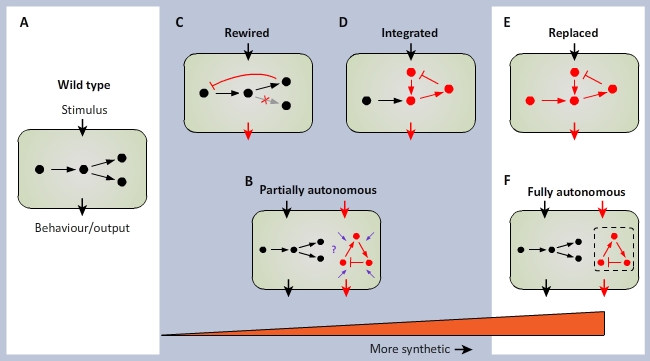 After Nandagopaland and Elowitz (2011). A continuum of synthetic biology. Wild-type cells (A) can be subject to two basic types of synthetic manipulation. (B) Autonomous synthetic circuits, consisting of ectopic components, may be introduced into the cell. Such circuits process inputs and implement functions (red arrows) seperate from the endogenous circuitry (black). However, unknown interactions with the host cell may affect their function (purple arrows) (C)An alternative is to rewire (red lines) the endogenous circuits themselves to have new connectivity. (D) Extending this line of synthetic manipulation, synthetic circuits could be integrated into appropriately rewired endogenous circuitry to act as sensors and to implent additional functionality. Ultimate goals of this program are to be able to design and construct (E) synthetic circuits that can functionality replace endogenous circuits or (F) fully autonomous circuits that operate independently of the cellular mileu. Nagarajan Nandagopal and Michael B. Elowitz. (2011). Synthetic Biology: Integrated Gene Circuits. SCIENCE, Vol. 333: 1244-1248. Another very important and necessary feature of biological parts is orthogonality, which means in this context that independent devices can be combined unrestrictedly. This principle derives from the area of engineering and aims to alter subsystems, without impeding others. This way cells can be modified as requested, resulting in a predictable behaviour. Among others, this technique can be beneficial for the effective production of certain substances, like biofuels or antibiotics.
|
|
| ↑ Back to top! |

|
 "
"