Team:Bielefeld-Germany/Results/thermo
From 2012.igem.org

Summary
Initially some trials of shaking flask cultivations were made with different parameters to identify the best conditions for the production of the His-tagged laccase LttH from [http://www.dsmz.de/catalogues/details/culture/DSM-7039.html?tx_dsmzreso Thermus thermophilus HB27] named TTHL. Due to the absence of enzyme activity of the enzyme in the cell lysate a purification method was established (using Ni-NTA-His tag resin). Using E. coli KRX containing BioBrick <partinfo>BBa_K863010</partinfo>, TTHL could not be detected by SDS-PAGE (molecular weight of 53 kDa) or by activity test. Therefore a new BioBrick <partinfo>BBa_K863012</partinfo> was constructed and expressed in E. coli Rosetta-Gami 2. With this expression system the TTHL could be detected by SDS-PAGE and purified by using a small scale Ni-NTA column. The fractionated samples were tested regarding their activity. TTHL was shown to oxidize ABTS. After measuring activity of TTHL a scale up of the fermentation was successfully implemented up to 6 L.
Contents |
Cultivation, Purification and SDS-PAGE
Shaking Flask Cultivation
The first trials to produce the LttH-laccase from [http://www.dsmz.de/catalogues/details/culture/DSM-7039.html?tx_dsmzresources_pi5 Thermo thermophilus HB27] (named TTHL) were performed in shaking flasks with various volumes (from 100 mL up to 1 L flasks, with and without baffles) and under different cultivation conditions. The best cultivation condition for <partinfo>BBa_K863010</partinfo> expressed in E. coli was screened by varying the temperature, the chloramphenicol concentration,induction strategy and cultivation time. Furthermore, E. coli was cultivated with and without 0.25 mM CuCl2 in the medium to provide a sufficient amount of copper, which is needed for bilding the active center. Under the screened conditions no biological active TTHL could be produced. Therefore another BioBrick was constructed and another chassi was chosen. To improve the expression another BioBrick <partinfo>BBa_K863012</partinfo> was used, which has a constitutive promoter instead of the T7 promoter system. Additionally, the strain E. coli Rosetta-Gami 2 was chosen, because of its ability to translate rare codons. TTHL was then produced under the following conditions:
- flask design: shaking flask without baffles
- medium: LB-Medium
- antibiotics: 60 µg mL-1 chloramphenicol and 300 µg mL-1 ampicillin
- temperature: 37 °C
- cultivation time: 24 h
The reproducibility of the measured data and results were investigated for the shaking flask cultivation, but not yet for the bioreactor cultivation.
Fermentation of E. coli KRX with <partinfo>BBa_K863012</partinfo>
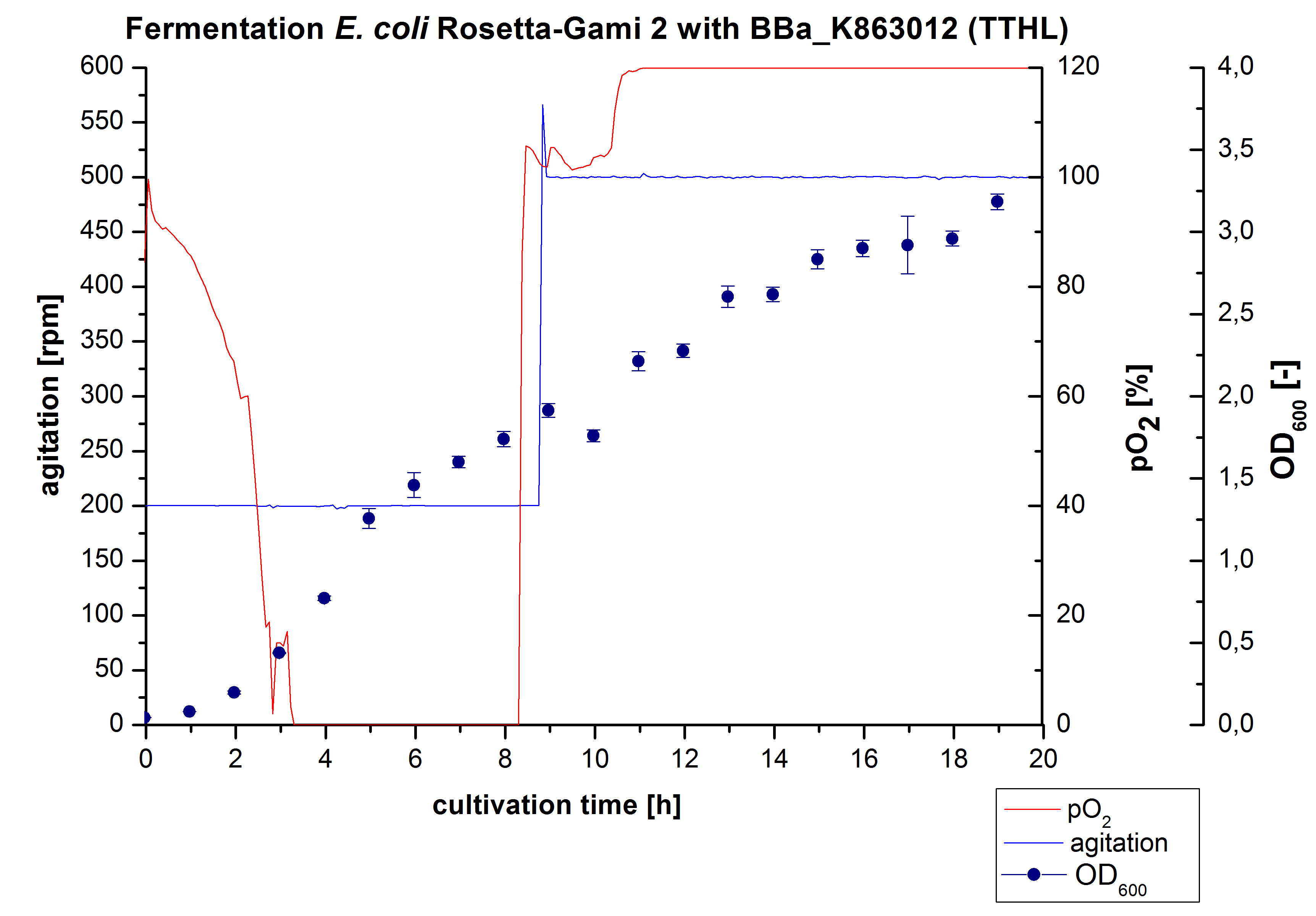
After measuring activity of TTHL we made a scale-up and cultivated E. coli Rosetta-Gami 2 expressing <partinfo>BBa_K863000</partinfo> in a Bioengineering NFL22 fermenter with a total volume of 6 L. Agitation speed, pO2 and OD600 were online monitored and are illustrated in Figure 1. No initial lag phase was noticeable. Due to the cell growth the pO2 decreased,breakdown of the control unit resulted in a drop to 0%. After a cultivation time of 9 hours the agitation speed was therefore increased manually up to 500 rpm, which resulted in a higher pO2 value of more than 100 % for the rest of the cultivation. During the whole process the OD600 increased slower compared to the fermentation of E. coli KRX expressing <partinfo>BBa_K863000</partinfo> or <partinfo>BBa_K863005</partinfo>. The maximal OD600 was reached after 19 hours cultivation time at which point the cells were harvested.
Purification of TTHL
The cells were harvested by centrifugation and resuspended in Ni-NTA-equilibrationbuffer, mechanically disrupted by high pressure homogenization and centrifuged. After preparing the cell paste the TTHL could not be purified with the 15 mL column, due to a not available column. For this reason a small scale purification (6 mL) of the supernatant of the homogenisation was made with a 1 mL Ni-NTA-column.
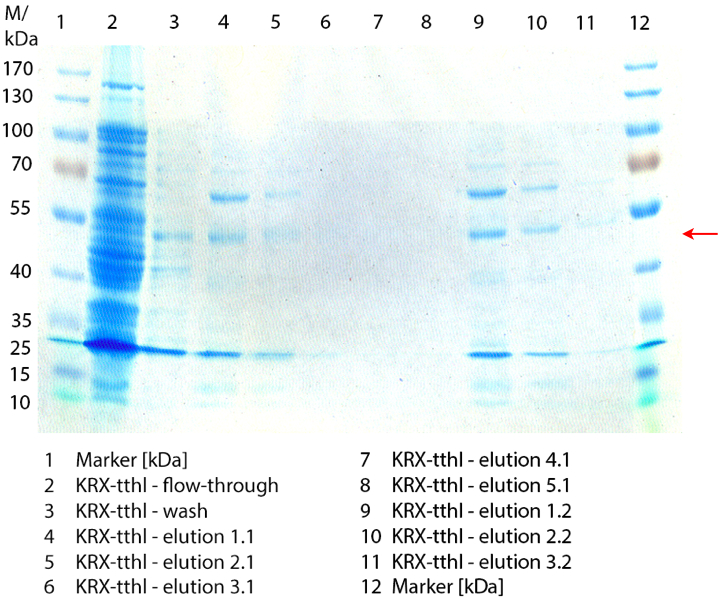
SDS-PAGE of purified TTHL
Figure 2 shows the SDS-PAGE of the purified E. coli Rosetta-Gami 2 lysates fermented in 6 L Bioengineering NFL22 fermenter. Additionally the flow-through, wash and all elution fractions (1 to 5) are shown. TTHL has a molecular weight of 53 kDa and the corresponding band is marked with a red arrow. The TTHL band can be found in fractions 1 to 3, but not in the other two elution fractions. Furthermore there are some other non-specific bands, which could not be identified. To improve the purification an 15 mL column was implemented.
Activity analysis of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 TTHL]
Initial activity tests of purified fractions
Before Regionals
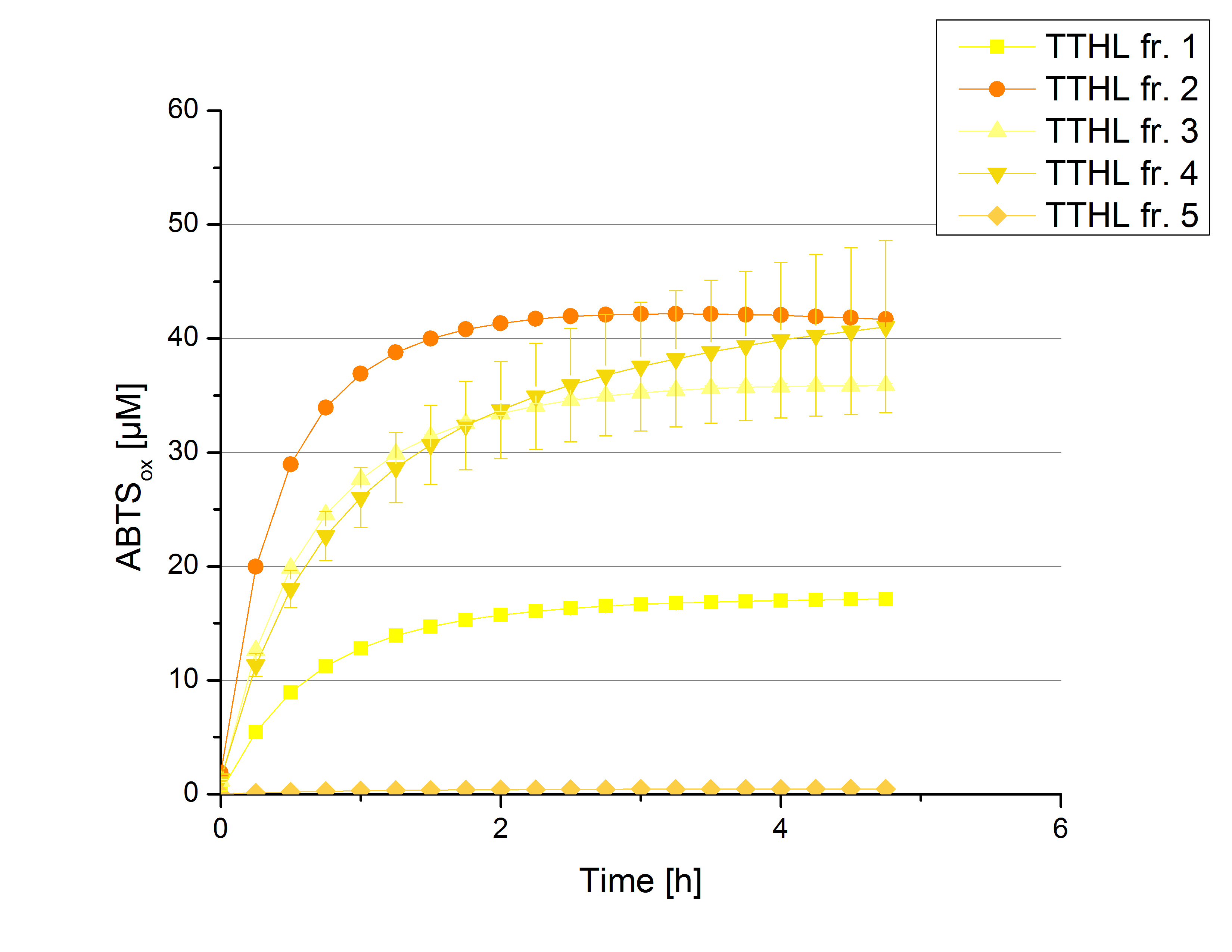
There was no activity measurable after cultivation and purification of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863022 BBa_K863022] under the control of a T7 promoter. Activity tests of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 TTHL] under a constitutive promoter did reveal TTHL laccases capable of oxidizing ABTS. Fractions 1 to 5 of the purification above were rebuffered with deionized H2O and incubated with 0.4 mM CuCl2 for 2 hours. Activity measurements were performed using 140 µL sample, 0.1 mM ABTS and 100 mM sodium acetate buffer (pH 5) to a final volume of 200 µL. The change in optical density at 420 nm was detected, reporting the oxidization of ABTS through laccases. Fractions 2 to 5 show activity (Figure 3). Fraction 2 seems to contain most of TTHL showing the highest activity compared to the other fractions: 40% of the used ABTS has been oxidized after 2 hours. Based on these results protein concentrations have to be determined and the activity of the TTHL laccase can be characterized in further experiments including pH optimum and activity in regard of temperature shifts.
Since Regionals
The purificated fractions of the cultivation after the Regional Jamborees in Amsterdam were tested concerning their protein concentration. After re-buffering the protein concentration was determined again and all fraction were incubated with 0.4 mM CuCl2. For the initial activity test
12L Fermentation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012]
Purification of TTHL
The harvested cells were resuspended in Ni-NTA- equilibration buffer, mechanically disrupted by homogenization and cell debris were removed by centrifugation and microfiltration via[http://www.millipore.com/catalogue/module/C7493 Millipore Pellicon XL 50]. The supernatant of the cell lysate was concentrated with [http://www.millipore.com/catalogue/module/C7493 Millipore Pellicon XL 50] with 10 kDa) and loaded on the Ni-NTA column (15 mL Ni-NTA resin) with a flow rate of 1 mL min-1 cm-2. Then the column was washed with 10 column volumes (CV) Ni-NTA equilibration buffer. The bound proteins were eluted by an increasing Ni-NTA elution buffer step elution from 5 % (equates to 25 mM imidazol) with a length of 50 mL, to 50 % (equates to 250 mM imidazol) with a length of 80 mL and finally to 100 % (equates to 500 mM imidazol) with a length of 80 mL. This strategy was chosen to improve the purification caused by a step by step increasing Ni-NTA-elution buffer concentration. The elution was collected in 10 mL fractions. Due to the high UV-detection signal of the loaded samples and to simplify the illustration of the detected product peak only the UV-detection signal of the wash step and the elution are shown. A typical chromatogram of purified laccases is illustrated here. The chromatogram of the TTHL elution is shown in Figure 5:
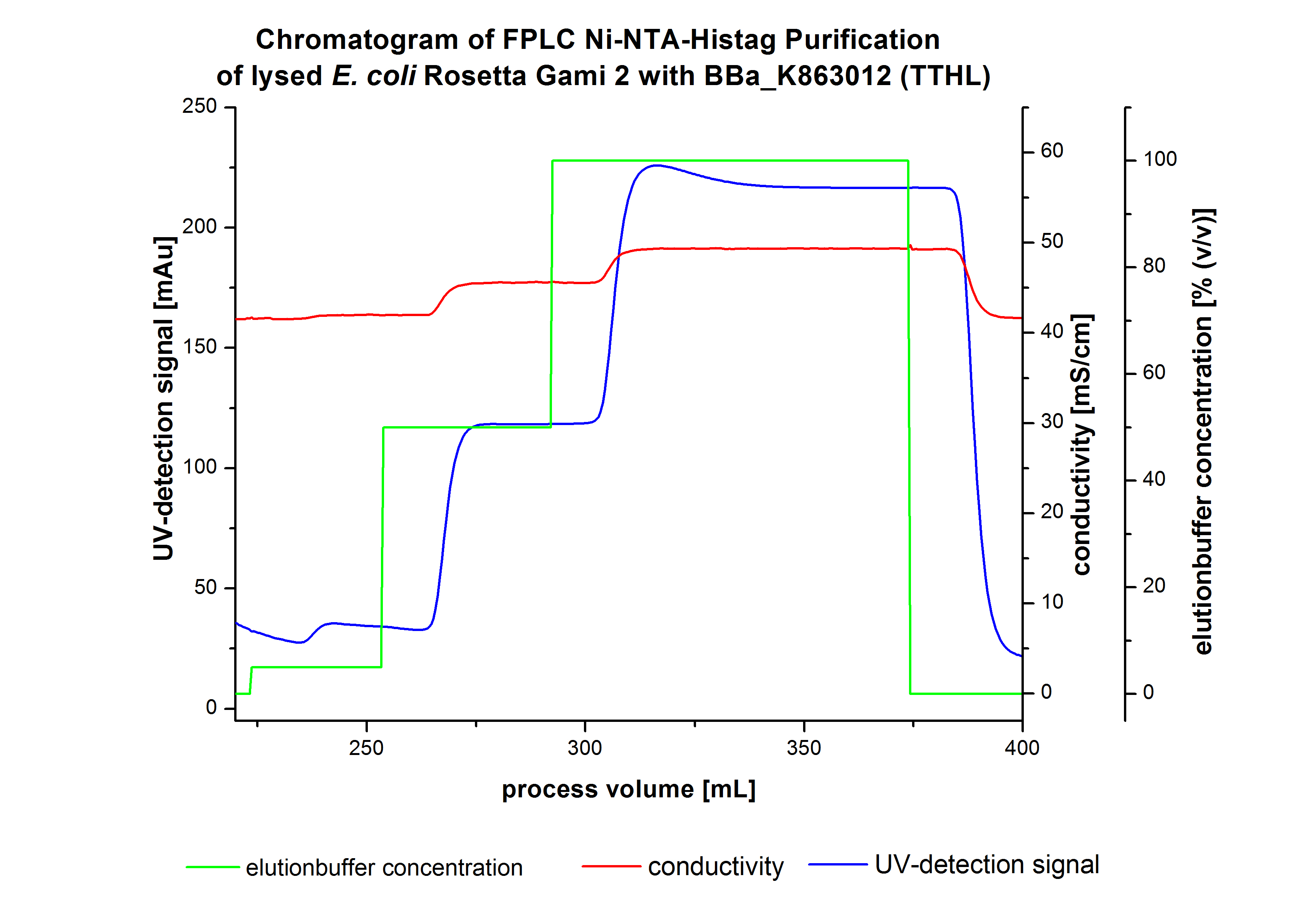
Contrary to our expectations, the chromatogram shows one distinguished peak. This peak was detected at a Ni-NTA-equilibration buffer concentration of 100 % (equates to 500 mM imidazol) and resulted from the elution of bounded proteins. Earlier measurements of other bacterial laccases showed that the elution of these laccases begins with a elutionbuffer concentration of 50 %(equates to 250 mM imidazol). One explanation of this result could be a low concentration of the produced TTHL. Just to be on the safe side all elution fractions were analyzed by SDS-PAGE to detect TTHL. There were no further peaks detectable. The following increasing UV detection signal by increasing concentration of the eltutionbuffer results from the rising imidazol concentration of the Ni-NTA elution buffer. The corresponding SDS-PAGES are shown in Figure 4.
| 55px | | | | | | | | | | |
/partinfo>. The maximal OD
 "
"