Team:Bielefeld-Germany/Labjournal/week4
From 2012.igem.org
(Difference between revisions)
(→Thursday May 24th) |
(→Wednesday May 23rd) |
||
| Line 33: | Line 33: | ||
*'''Team Bacterial Laccases''': | *'''Team Bacterial Laccases''': | ||
| - | ** After there were no colonies on our pSB1C3 + | + | ** After there were no colonies on our pSB1C3 + xccl (''Xanthomonas campestris'') transformation plate we did the transformation with the same ligation preparation again. The other ligation with pSB1C3 + ecol (''E. coli'') showed colonies so we started colony PCRs to find positive colonies. Sadly the colony PCRs showed no products but the problem was that we just had the long overhang primers (E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS ). Therefore we ordered the F and R [https://2012.igem.org/Team:Bielefeld-Germany/Protocols#Primers primers]. |
===Thursday May 24th=== | ===Thursday May 24th=== | ||
Revision as of 15:36, 22 September 2012
Contents |
Week 4 (05/21 - 05/27/12)
weekly seminar:
- Lab service: Isabel
- We try to establish a collaboration with the iGEM team from SDU-Denmark
- Got our distribution kits
- First successful cloning and cultivations
- Who wants to be a summer school teacher?
- We will not travel to the ACHEMA because only local teams are invited
- Do we want to participate in the Biolympics? (It's a sports party with fun organized by the [http://bts-ev.de/ bts])
Monday May 21st
- Team Modeling: Our aims for modeling:
- model the expression of the laccases in the organisms.
- model the activity of our enzymes.
- model the interesting parts of a clarification plant (the part witch are interesting for our cleaner.
- Team Bacterial Laccases:
- We wanted to clone our laccase PCR products xccl and ecol in pSB1C3 backbone. Therefore we did some restriction digests on the PCR products and the vector [http://partsregistry.org/Part:BBa_J04450 BBa_J04450].
Tuesday May 22nd
- Team Bacterial Laccases:
- Ligation of the digested PCR products in pSB1C3 backbone and transformation in KRX electro-competent cells.
Wednesday May 23rd
- Team Student Academy:
- Repetition of the transformation of [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 BBa_I13522] with new competent E. coli KRX cells. Got intense red fluorescing colonies but no green fluorescing colonies. Made a backup of E. coli KRX [http://partsregistry.org/Part:BBa_J04450 BBa_J04450].
- Asking for other plasmids containing GFP at the working groups of our University.
- Team Bacterial Laccases:
- After there were no colonies on our pSB1C3 + xccl (Xanthomonas campestris) transformation plate we did the transformation with the same ligation preparation again. The other ligation with pSB1C3 + ecol (E. coli) showed colonies so we started colony PCRs to find positive colonies. Sadly the colony PCRs showed no products but the problem was that we just had the long overhang primers (E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS ). Therefore we ordered the F and R primers.
Thursday May 24th
- Team Student Academy:
- Made a liquid culture of E. coli KRX with [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] at 30 °C. There was no fluorescence.
- Transformation of [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] into E. coli KRX. Also got intense red fluorescing colonies.
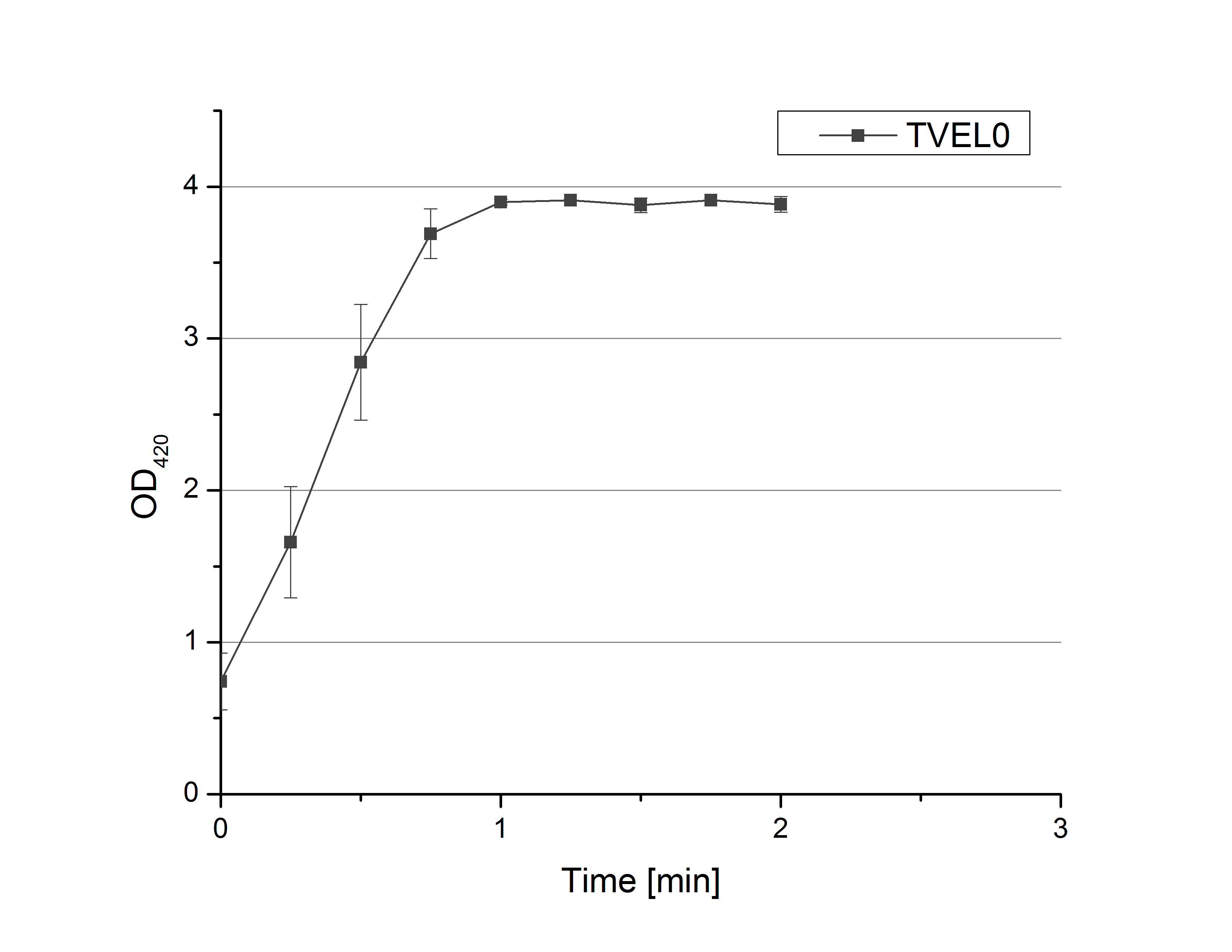
- Team Activity Tests: On our schedule today was testing our bought laccases and improving our protocol in case it won't work. We got familiar with the flat bottom microplates and got a briefing into using the Tecan correctly. We started with the following setup: 100 mM sodium acetate (pH 5), 5 mM ABTS, 8 U TVEL0 laccase, ad 200 µL with deionized water. Then we measured the absorption at 420 nm every 15 seconds over a time period of 2 minutes. Immediately our samples turned dark blue but unfortunately the changeover was out of range to be detected by Tecan. With this information we needed to reduce the laccase concentration to get measurable results. We have chosen to try another measurement with 0.1 U TVEL0 laccase and it worked! The result was a nice saturation curve but it reached a too high, because not good measurable, optimum within 1 minute (Fig. 1). To avoid the loss of important data in the beginning of the reaction and to reduce the saturation OD we decided to slow everything down by using less ABTS.
Friday May 25th
- Team Bacterial Laccases:
- We had to do the PCR on T. thermo laccase again because after the purification of the last PCR product the DNA amount was very low.
Saturday May 26th
Sunday May 27th
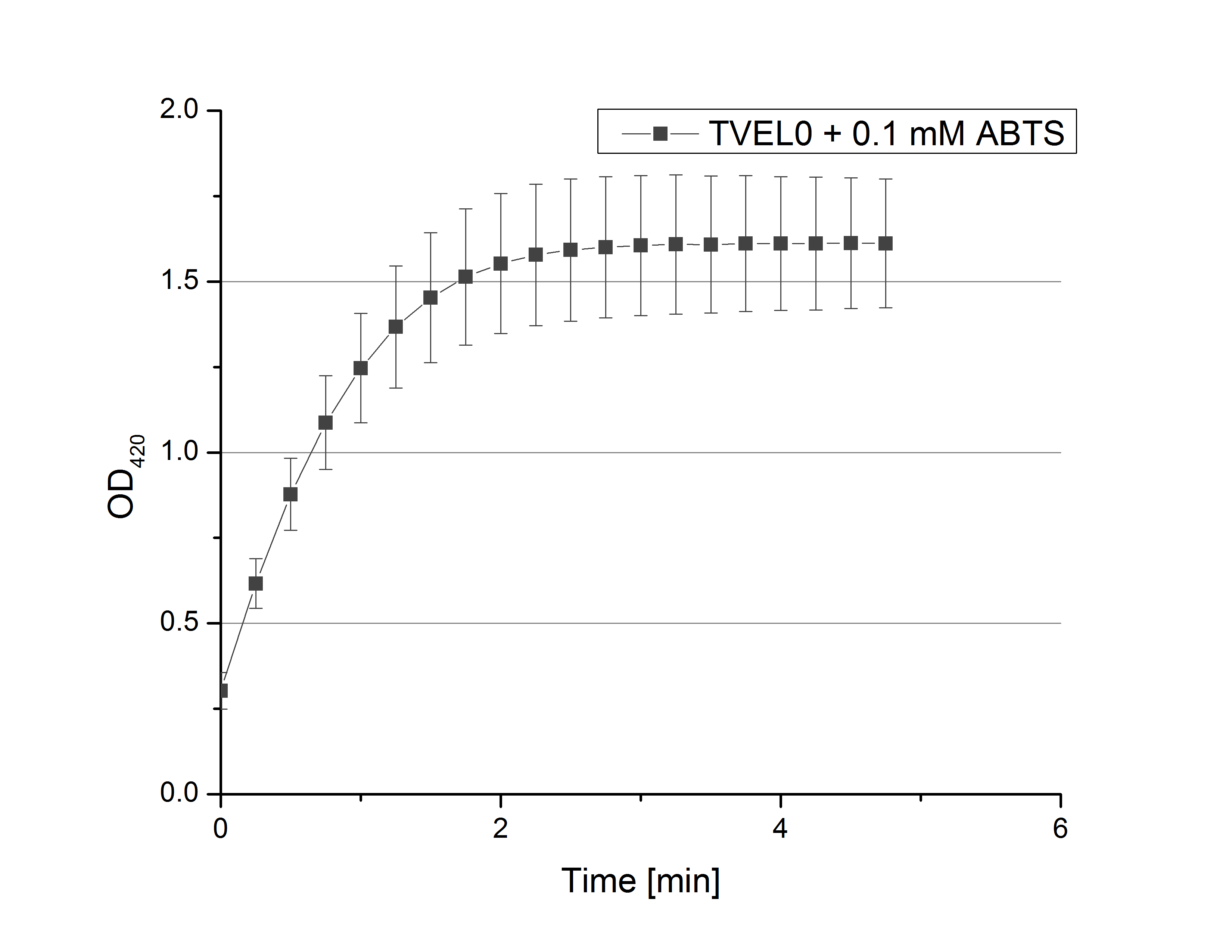
- Team Activity Test: Remember our first activity measurements? The activity was awesome and reached quickly its saturation. So today we decided to have a lazy Sunday and reduce absorption maximum and reaction speed, too. For this we used 0.1 mM ABTS instead of 5 mM ABTS and we got good results. The saturation reached its maximum at a OD420 of ~1.6 after roundabout 2 minutes (see Fig. 2). Taking together we halved the maximal OD420 and doubled the reaction time until maximum is reached with using 0.1 mM ABTS instead of 5 mM ABTS.
| 55px | | | | | | | | | | |
 "
"