Team:Bielefeld-Germany/Labjournal/week13
From 2012.igem.org
Contents |
Week 13 (07/23 - 07/29/12)
- From 07/23 - 07/25/12, some of our team members participated at the CAS conference in Munich.
Monday July 23rd
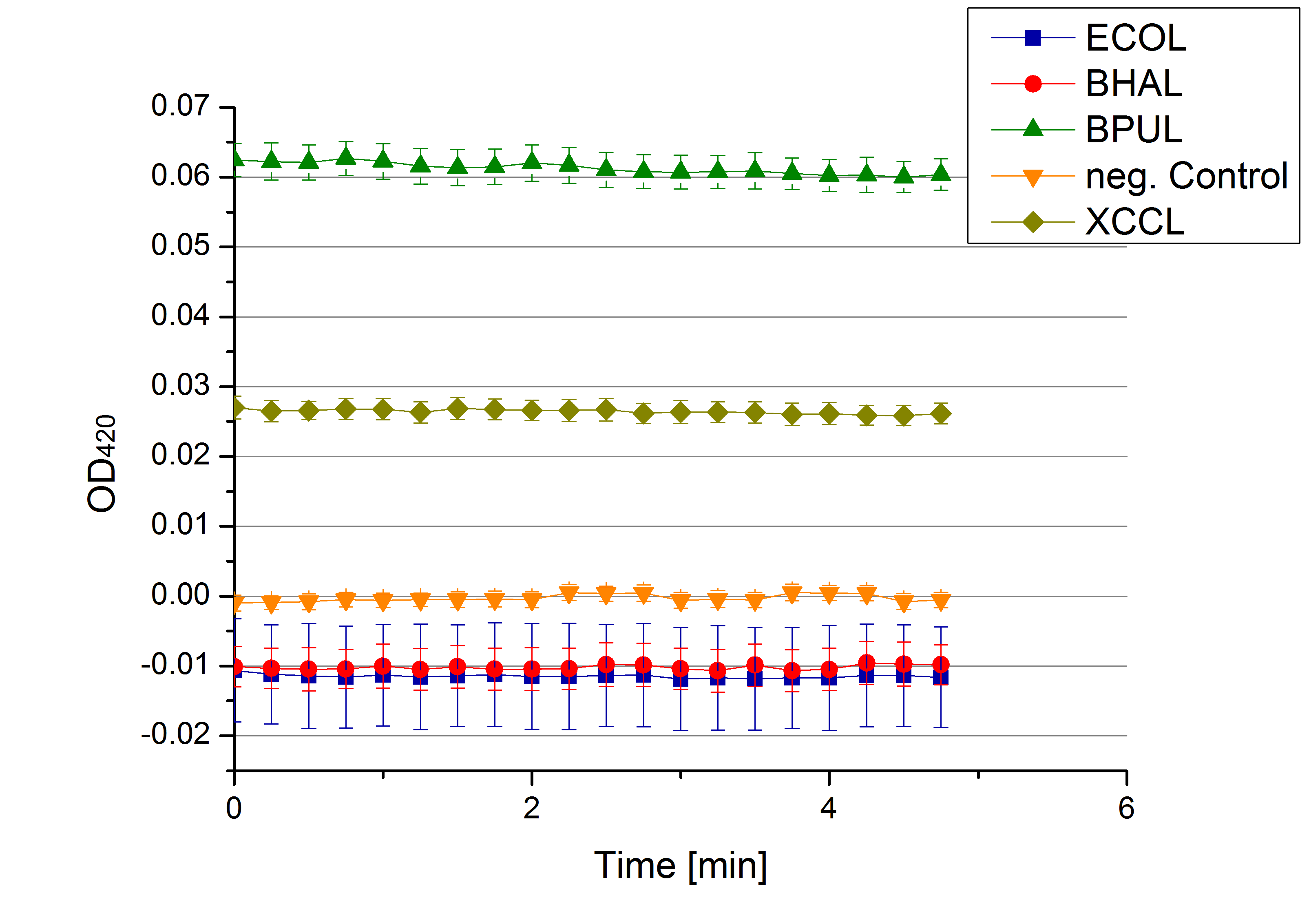
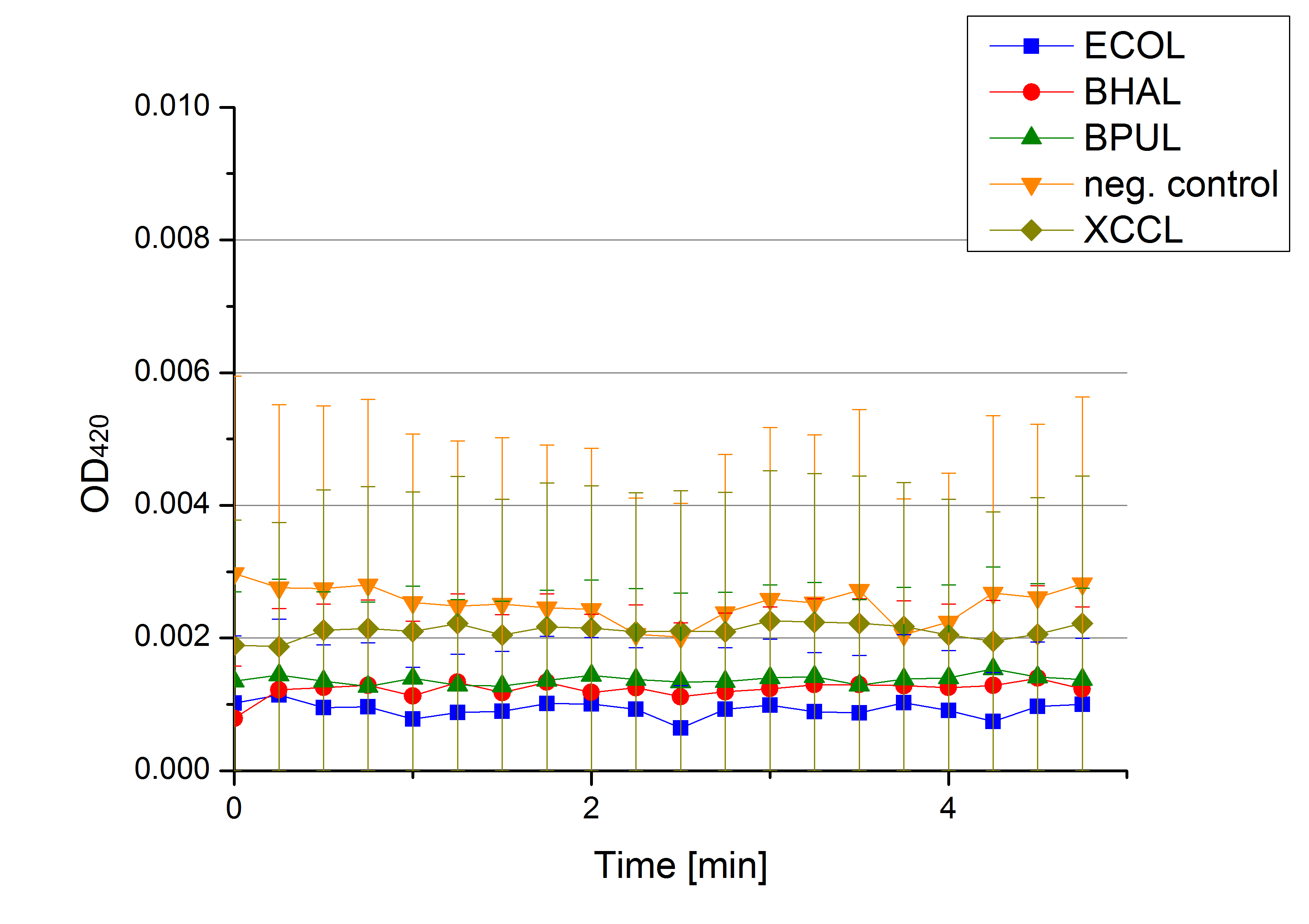
- Team Activity Tests: Today was the moment of truth. We received the first recombinant laccases from the cultivation team. We started with testing the supernatant from the cultivated cells to check whether they secrete laccase. We added 0,1 mM ABTS in each well that was filled with supernatant from E.coli KRX that contained the plasmid to produce ECOL, BPUL, BHAL, XCCL or E. coli KRX (negative control) and measured as usual. There was no change in the OD so that we assume that no secretion takes place. Afterwards we tested potential laccase activity by filling each well with laccase and buffer and added 0,1 mM ABTS. Unfortunately no activity was seen. We felt like we could not take the laziness of our laccases. We thought and discussed what the reason could be for them to be inactive.
- Is there something wrong with the transformed construct, maybe a mutation?
- Was the laccase synthesized but is inactive for some reason?
- Was the laccase produced but not in a high enough amount so that we can´t detect its activity in the first place?
Another thought was that there might be copper missing. Since laccase is a copper-enzyme it might be necessary to add copper to the medium whenever it is produced in such high amounts. It´s late, so we will do that tomorrow.
- Team Bacterial Laccase: The transformation from the 21st July showed no results.
- Team Fungal Laccases: Successful PCRs of laccase gene Tv5 from Trametes versicolor with plasmid DNA from Uni Greifswald as template.
- Team Site Directed Mutagenesis: Transfered 4 colonys of each transformation to an new dish for plasmid-isolation
Tuesday July 24th
- Team Site Directed Mutagenesis: Plasmid-isolation of all 16 different colony-dishes
- Team Activity Test: Today is copper day! First we divided each sample (those from yesterday) and added 2mM or 4mM to the halfs, respectively. We incubated our samples for 2h and hoped that the concentration gradient would cause the laccases to exchange the ion they have integrated instead against copper. We measured their activity again after those 2h but there was no change seen. Team Cultivation plans to add copper from the very beginning of cultivation next time.
- Team A. thaliana laccase: Since our cDNA is now ready to go and also our primers already arrived we started a PCR. Check protocols for more information about the exact setup. Unfortunately no bands could be seen via gel electrophoresis. We blamed the high salt concentration and decided that a some cleaning via ethanol precipitation might be a solution to our problem.
Wednesday July 25th
- Team Site Directed Mutagenesis: Test restriction (with the enzym of the mutated restriction-site) of the Plasmids from the 16 different colonies revealed that two bpul, one ecol and two tthl mutants have the right bands in the gel and seem be mutated correctly. xccl-colonies are all unmutated (no Band at 3636).
- SDM-PCR with the two B.pumilus III(III)+IV(IV) as template using the SDM-primermix to mutate bp 2317
- SDM-PCR with X.campestris with the 2247-primermixand another one with the primermix to mutate 3633
- Prepared test restriction-positives for Sequencing
- Team Cultivation & Purification:
- We made precultures of E.coli KRX without plasmid and with BBa_K863005 , pBpL6 as well as with laccases from B.halodurans or X.campestris.
Thursday July 26th
- Team Modeling and Team Sponsoring: meeting Mr. ??? from the clarification plant of Schloß Holte and finding a new cooperation partner. He want to give us the information we need to equate our model and design our cleaner. On top he wants to ask if the clarification plant can sponsor our project.
- Team Site Directed Mutagenesis: Went on with the PCR products and transformed them into XL1 Blue
- Team Fungal Laccases: We did PCR on Trametes versicolor laccase Tv5 and Pycnoporus cinnabarinus Pc35 with Tv_lac10.P.FW / Tv_lac10.S.RV and Pc_lac35.P.FW / Pc_lac35.S.RV primers. Additionally we want the laccases with overhanging ends for cloning in our shuttle vector for expression in Pichia pastoris. Therefore we used Pc_lac35_FW_oS / Pc_lac35_RV and Tv_lac5_FW_oS / Tv_lac5_RV primer pairs.
- Team Cultivation & Purification:
- Glycerine cultures were made of all given BioBricks, to use them for precultures.
- Made new precultures analogous to 07/25.
- Found out, that it could be important to add CuCl2 to the medium, because the copper is important for the active center of the laccases. As an alternative we could incubate them with copper before measuring the activity.
Friday July 27th
- Team Site Directed Mutagenesis: picked colonies and plated them for plasmidisolation.
- Team Cellulose Binding Domain: After a chat with teammates made the decision to only use Cellolose binding domains and not carbohydrate binding domains to keep it comparable. This means we won't use the binding domains of Bacillus halodurans.
- Redesigned Primers for CBDclos - added the bases TA in between RBS and ATG since the RBS can be made stronger by adding more adenines in the sequence upstream of the RBS
- Made similar primers for CBDcex (the CBD of the Cellulomonas fimi exoglucanase we got for the fermentationgroup)
- Checked the Osaka_2010 page about their BBa_K392014 BioBrick which should be a binding motif but is the Glycosyl hydrolase domain. I guess them messed it up...
- Team Fungal Laccases: The PCR on Tv5 laccase was positive after the first trial for both primer pairs. For the Tv35 laccase the PCR didn’t work.
- Team Cultivation & Purification:
- We cultivated E.coli KRX with BBa_K863005, laccases from B.halorudans, X.campestris as well as pBpL6. As negative control we used E.coli KRX.
- --> Settings: 300mL flasks without baffles, final volume: 60mL, autoinduction medium, 0,25mM CuCl2, 30°C
- We made glycerine cultures of homologous culture of E.coli KRX with pBpL6.
Saturday July 28th
- Team Activity Tests: See, we are back already! Today we received different samples from Escherichia coli, Bacillus pumilus, Bacillus halodurans C-125, Thermus thermophilus, Xanthomonas campestris pv. campestris B100 and E. coli KRX (negative control - cells without plasmid). The cells were all cultivated at 30°C and supplied with copper. We tested each one of them but none of them showed laccase activity.
- Team Fungal Laccases: Purification of Trametes versicolor Tv5 laccase.
- Team Cultivation & Purification:
- Cells of cultivation 07/27 were disrupted via sonification and given to the activity test team.
Sunday July 29th
- Team Cultivation & Purification:
- Preparation of all the stuff needed for SDS-Pages.
- Made a new preculture of E.coli KRX without plasmid and with BBa_K863005, laccases from B.halodurans, X.campestris, as well as pBpL6 and a new construct of T.thermophilus.
| 55px | | | | | | | | | | |
 "
"