Team:Bielefeld-Germany/Project/Database
From 2012.igem.org
Contents |
One important thing while working in such a big team like ours is the organization of samples. Therefore we designed our own database in which we can make entries for each sample we use. This page gives you detailed information on how it was designed and implementet.
Design of the database
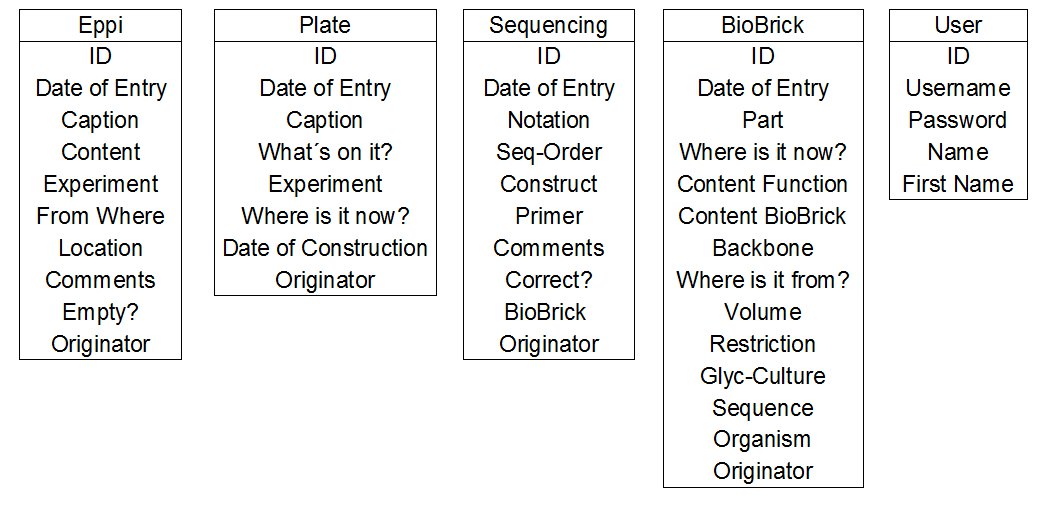
- initial design : First of all we considered which tables and which listings of the different tables were necessary. Here you can see our first abstract thoughts. This part of the design of a database is called the initial design of a database.
We need the User table for the login and for the allocation of entries to the team members. In the User table we create an ID specific for each team member. This ID can be used as a foreign key in the other tables (see figure 1).
Additionally we want to realize the following particularities:
- For the Eppis table (and with Eppi we always mean reaction tubes) it is very important, that you can always find out were a sample is. So we decided that we have to write a script which controls if you insert a place of storage to your database entry, otherwise this entry could not be saved.
- For the Plates table we need a script which checks if the date of construction is older than two weeks. If the entry is older than these two weeks, the entry will be marked in red automatically, thus the skript provides an easy way to check if all our plates are fresh.
- For the BioBricks table we want to name our BioBricks automatically. Therefore a script should be implemented, which assignes a BioBrick number, according to the affiliation to either eukaryotic, bacterial, fungal or others.
- Another very important aspect was, that the editing of entries is secured against mistakes, for example one person creating an entry and and another person deletes it by mistake. Hence, we need a script which conserves the entries. It saves the entry as a copy in the datebase but just shows the newest copy of an entry. With this script it is possible to rebuild an entry.
- We did the same for the editing script.
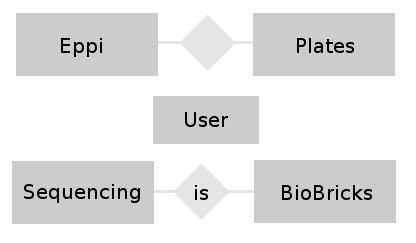
- conceptual design: In this phase of the design we try to find relations between the different tables and build a entity-relationship-model. This model helps to visualize the entities (our different tables), their attributes (the entries belonging to a table) and the relationship between these entities.
- We want to combine the Eppi table with the Plates table and the BioBrick table with the Sequencing table.
- The User table is combined to all tables because of the foreign keys (see figure 2).
- logical design : After this we thought about the logical design of our database. In this phase the conceptual design is implemented with the help of a precise data model, for example a relational design. We decided to use such a design.
Relational design of the Eppi table (exemplary):
| Eppi ( | ID INT(11), |
| DataID INT(11), | |
| Caption TEXT, | |
| Content TEXT, | |
| Experiment VARCHAR(255), | |
| Where2 VARCHAR(255) , | |
| Location TEXT, | |
| Comments TEXT, | |
| Empty ENUM ('N', 'J'), | |
| Create_User INT(11), | |
| Create_Date DATETIME, | |
| Edit_Date DATETIME, | |
| Delete ENUM ('N', 'J') | |
| Delete_User INT(11), | |
| Delete_Date DATETIME ) |
Programming the Database
Used Languages
As programming language we used PHP. PHP is a general-purpose server-side scripting language and was originally designed for web development to produce dynamic web pages. It is one of the first developed server-side scripting languages to be embedded into an HTML source document and can be used with many relational database management systems (RDBMS). As RDBMS we used MySQL for the construction of the tables in the database. MySQL is the world's most used open source RDBMS for the use in web applications. The abbreviation SQL stands for Structured Query Language. For the optical design of the tables we used HTML. HTML stands for HyperText Markup Language which is the main markup language for displaying web pages and other information that can be displayed in a web browser. We used Cascading Style Sheets (CSS) in order to not have to design each table individually. CSS is a style sheet language used for describing the look and formatting of a document written in a markup language like HTML.
Realization of the design
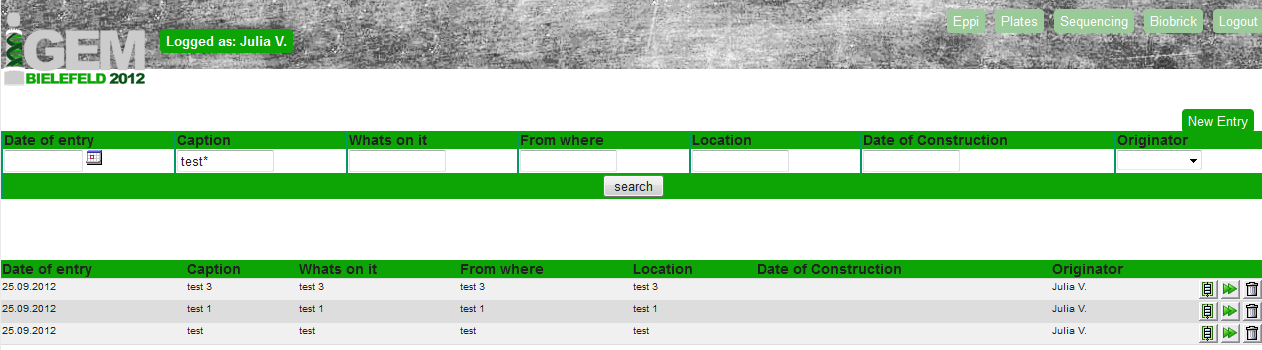
For the creation of an database entry it is necessary to be logged in as an user with an User name and a password (both saved in the User table).
After the successful log in it is possible to edit the Eppi, the Plates, the Sequencing or the BioBrick table. The User table is only visible for the administrator.
A search tool was implemented (see upper part of figure 3). With using the underscore ( _ ) for one letter after the search text and the asterisk ( * )for any number of letters after the search text you can look for specific entries in the selected table.
Important Functions
Here we listed some important scripts we used. (click to the black arrows for the next one)
Using our database - Downloads
We wanted to provide our database with all other iGEM Teams. Therefore we tryed to upload the program to the igem.org page. But we have the problem, that the program together with a Server2Go is too big for uploading to the iGEM server, so we just had the opportunity to upload it to our own server. The Server2Go is a web server that runs out of the box without any installation and on write protected media. This means that a web application based on Server2Go can be used directly from the download and so we have the possibility to provide you with the complete database with all its functions. Nevertheless we uplaoded a .zip file with the whole source code on the iGEM server. So you can choose between two different downloads.
- The first download only includes the scripts, the php, the css and the html files. Without the webserver you can not use the database on its own. This download is only for for unterstanding of the methods and programming techniques.
Media:iGEM_bielefeld_database_scripts.zip
- The second download is the complete program and can be run without any further software. After downloading, you just need to unzip the file, start the Start.exe and the database is automaticlly opened in your web browser.
[http://2012.igem-bielefeld.de/database/iGEM_bielefeld_database.zip iGEM_bielefeld_database.zip]
| 55px | | | | | | | | | | |
 "
"