Team:Bielefeld-Germany/Protocols/Production
From 2012.igem.org
KevinJarosch (Talk | contribs) (→ÄKTA method) |
(→Syringe method) |
||
| (12 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| + | <div id=page-title> | ||
| + | <span id=page-title-text> | ||
| + | Production | ||
| + | </span> | ||
| + | </div> | ||
<div id="grey_bg"> | <div id="grey_bg"> | ||
</html> | </html> | ||
| - | + | ||
=Production= | =Production= | ||
| - | + | ||
Here all our methods according to cultivation and purification are listed. | Here all our methods according to cultivation and purification are listed. | ||
| - | + | ||
==Pre-Cultivation== | ==Pre-Cultivation== | ||
| - | + | ||
===Precultivation of ''E.Coli'' KRX (with or without BioBrick)=== | ===Precultivation of ''E.Coli'' KRX (with or without BioBrick)=== | ||
* 50 mL [https://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB]-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs | * 50 mL [https://2012.igem.org/wiki/index.php?title=Team:Bielefeld-Germany/Protocols/Materials#LB_medium LB]-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs | ||
| Line 38: | Line 43: | ||
* Shaking at 140 rpm | * Shaking at 140 rpm | ||
| - | + | ||
==Cultivation== | ==Cultivation== | ||
| - | + | Cultivations in Autoinduction-Medium were supplemented with 0.1 % L-rhamnose as inducer, when the designated OD600 was reached, as described in the [http://www.promega.com/~/media/files/resources/promega%20notes/98/krx%20autoinduction%20protocol-%20a%20convenient%20method%20for%20protein%20expression.pdf?la=en Promega protocol] | |
===Expression of Laccase=== | ===Expression of Laccase=== | ||
| Line 71: | Line 76: | ||
* Harvest after 12-13 h | * Harvest after 12-13 h | ||
| - | |||
==Cell Harvesting== | ==Cell Harvesting== | ||
| - | + | ||
* Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C | * Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C | ||
| Line 79: | Line 83: | ||
* Resuspend the pellet in 5 mL [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#special_buffers_for_resuspending_the_cell_pellet special buffer] or [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography binding buffer] for each gramm of cell paste | * Resuspend the pellet in 5 mL [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#special_buffers_for_resuspending_the_cell_pellet special buffer] or [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Buffers_for_His-Tag_affinity_chromatography binding buffer] for each gramm of cell paste | ||
| - | + | ||
==Solubilization of inclusion bodies== | ==Solubilization of inclusion bodies== | ||
| - | + | ||
* centrifugation of the celllysate at 40,000 g for 30 minutes | * centrifugation of the celllysate at 40,000 g for 30 minutes | ||
| Line 89: | Line 93: | ||
* resuspend the pellet in SDS running buffer | * resuspend the pellet in SDS running buffer | ||
| - | + | ||
=Cell disruption strategies= | =Cell disruption strategies= | ||
| - | + | ||
==B-PER lysis (chemical lysis)== | ==B-PER lysis (chemical lysis)== | ||
| Line 126: | Line 130: | ||
The lysate is ready for a following purification step. | The lysate is ready for a following purification step. | ||
| - | + | ||
==Mechanical lysis== | ==Mechanical lysis== | ||
| - | + | ||
The method of choice to disrupt the cells depends on the amount of biomass. | The method of choice to disrupt the cells depends on the amount of biomass. | ||
| - | + | ||
===Mechanical lysis of the (shaking flask) cultivation=== | ===Mechanical lysis of the (shaking flask) cultivation=== | ||
| - | |||
| - | |||
| - | * | + | ====Sonication==== |
| + | |||
| + | * Sonication of the re-suspended pellet on ice | ||
** cycle number depends on the volume of the resuspended cells (e.g. 3 mL means 3 cycles) | ** cycle number depends on the volume of the resuspended cells (e.g. 3 mL means 3 cycles) | ||
** one cycle means sonification treatment for 1,5 min with Sonifier 450 by Branson, max. 50 %, cooled on ice, make sure not to heat the cells too much | ** one cycle means sonification treatment for 1,5 min with Sonifier 450 by Branson, max. 50 %, cooled on ice, make sure not to heat the cells too much | ||
| Line 147: | Line 151: | ||
**fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes) | **fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes) | ||
**homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles. | **homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles. | ||
| - | + | ||
===Mechanical lysis of the (bio-reactor) cultivation=== | ===Mechanical lysis of the (bio-reactor) cultivation=== | ||
| - | + | ||
====Cell disruption with a high-pressure homogenizer==== | ====Cell disruption with a high-pressure homogenizer==== | ||
| Line 156: | Line 160: | ||
* disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much | * disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much | ||
| - | + | ||
=Purification= | =Purification= | ||
| - | + | ||
| + | ===Ultra-/Diafiltration=== | ||
| + | <html> | ||
| + | <div style="float:right; width:400px; text-align:center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/5/5f/Bielefeld-Germany2011-filtrationmodule.jpeg" width="90%" height="90%" /> | ||
| + | <br/> | ||
| + | </div> | ||
| + | </html> | ||
| + | * Arrange the filtration module as shown on the right side. | ||
| + | * Microfiltration (0.22 µm) or cross flow filtration with 300 kDa (we used a Milipore Pellicon XL 300) membrane of sample before ultrafiltration. | ||
| + | * For concentrating the sample just filter it until the desired volume is left in the feed reservoir. For diafiltration (e.g. buffer exchange, desalting) dilute the feed reservoir several times and filter continously. | ||
| + | * Used membranes: [http://www.millipore.com/catalogue/module/C7493 Milipore Pellicon XL 50] or XL 100 membranes | ||
| + | ** 50 kDa cut-off | ||
| + | ** 50 cm<sup>2</sup> filtration area | ||
| + | ** tangential flow filter | ||
| + | ** Hydrophilic polyvinylidene fluoride membrane | ||
| + | * Used pump: SciLog TANDEM 1081 peristaltic pump | ||
| + | ** flow rate during filtration: 40 mL min<sup>-1</sup> | ||
| + | [[File:Bielefeld2012_IGEM_Diafiltration.jpg|thumb|center|500px| Filtrationsystem to purify the disrupted cells from cell debris, RED ARROW: Filtrationunits [http://www.millipore.com/catalogue/module/C7493 Millipore Pellicon XL 50] with 50 kDa (right) and 300 kDa (left)]] | ||
==His-tag affinity chromatography== | ==His-tag affinity chromatography== | ||
| Line 180: | Line 202: | ||
* Column: 1 mL HisTrap FF crude by GE Healthcare | * Column: 1 mL HisTrap FF crude by GE Healthcare | ||
| - | * Equilibrate with binding buffer(10mL) | + | * Equilibrate with binding buffer (10mL) |
| - | * Load sample onto column(max. 6 mL) | + | * Load sample onto column (max. 6 mL) |
* Wash with 10 mL binding buffer | * Wash with 10 mL binding buffer | ||
| - | * Elute with 5 mL of elution buffer | + | * Elute with 5 mL of elution buffer (500 mM imidazole) |
* Collect the eluate in 1 mL fractions, the purified protein is most likely in the first or second fraction | * Collect the eluate in 1 mL fractions, the purified protein is most likely in the first or second fraction | ||
* Re-equilibrate the column with binding buffer | * Re-equilibrate the column with binding buffer | ||
| Line 210: | Line 232: | ||
* Collect the eluate in 10 mL fractions | * Collect the eluate in 10 mL fractions | ||
* Elute remaining proteins with 100% Elutionbuffer (4 CV) | * Elute remaining proteins with 100% Elutionbuffer (4 CV) | ||
| - | [[File:Bielefeld2012 Chromatogram examplegrafik.jpg| | + | <br style="clear: both" /> |
| + | |||
| + | |||
| + | An typical chromatogram of purified laccases is illustrated in the following grafic: | ||
| + | |||
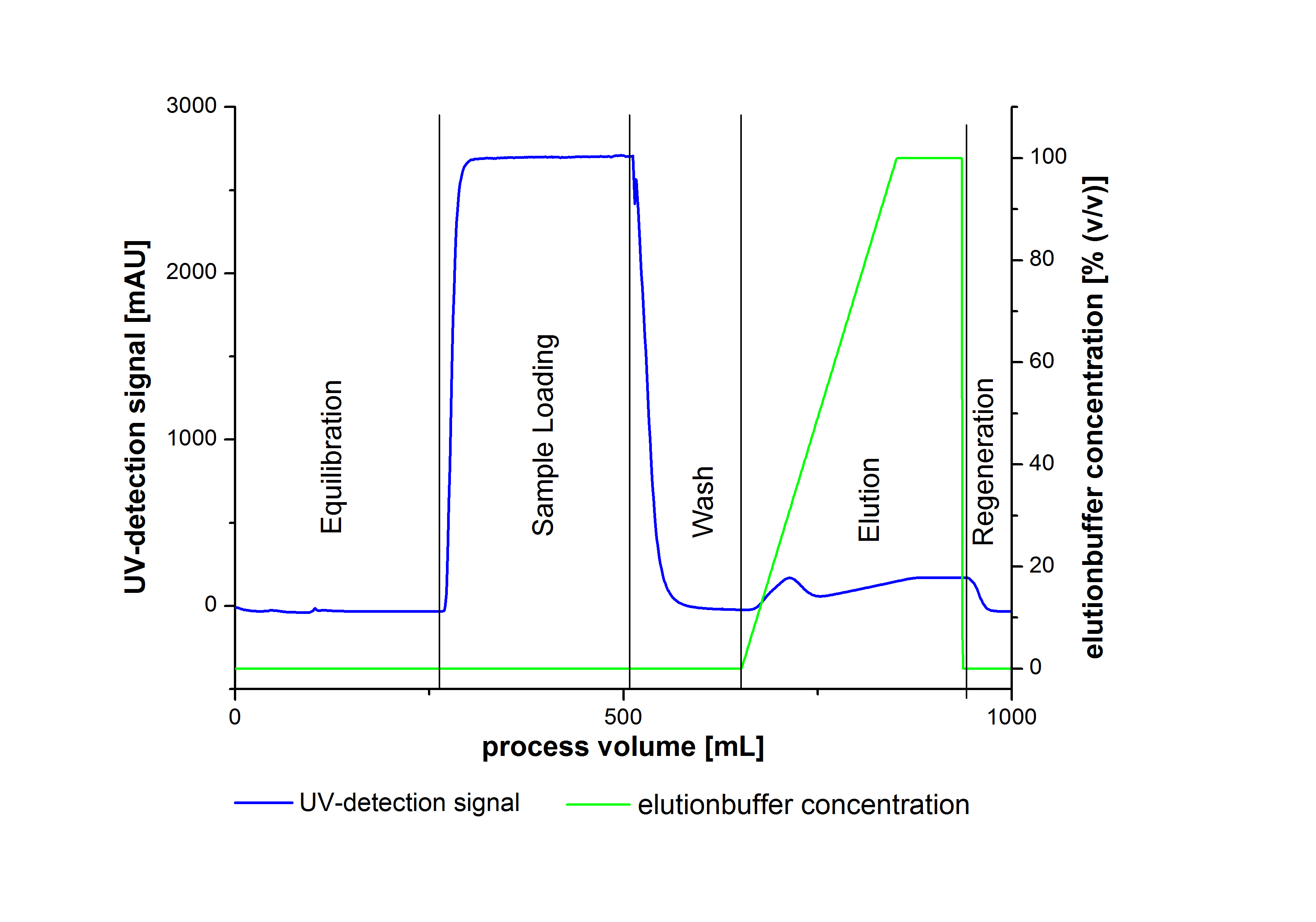
| + | [[File:Bielefeld2012 Chromatogram examplegrafik.jpg|800px|thumb|center| A typical chromatogram of the purification procedure of laccases. the different steps of the purification are illustrated: | ||
1.) Equlibration, 2.) Sample Loading, 3.) Wasch, 4.)Elution, 5.) Regeneration ]] | 1.) Equlibration, 2.) Sample Loading, 3.) Wasch, 4.)Elution, 5.) Regeneration ]] | ||
Latest revision as of 01:28, 27 October 2012
Contents |
Production
Here all our methods according to cultivation and purification are listed.
Pre-Cultivation
Precultivation of E.Coli KRX (with or without BioBrick)
- 50 mL LB-medium, if nessassary with 20-60 mg L-1 chloramphenicol or with 100-300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 37 °C
- Shaking at 140 rpm
Precultivation of E.Coli Rosetta Gami 2 (with or without BioBrick)
- 50 mL LB-medium with 60 mg L-1 chloramphenicol and with 300 mg L-1 ampicillin, in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 37 °C
- Shaking at 140 rpm
Precultivation of Pichia Pastoris GS115 (complex-medium)
- 50 mL YPD-medium in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 30 °C
- Shaking at 140 rpm
Precultivation of Pichia Pastoris GS115 (minimal-medium)
- 50 mL YNB-medium in 300 mL shaking flask with baffles (Schott) with silicon plugs
- 1 mL Glycerinculture or 1 colony
- Cultivation temperature: 30 °C
- Shaking at 140 rpm
Cultivation
Cultivations in Autoinduction-Medium were supplemented with 0.1 % L-rhamnose as inducer, when the designated OD600 was reached, as described in the [http://www.promega.com/~/media/files/resources/promega%20notes/98/krx%20autoinduction%20protocol-%20a%20convenient%20method%20for%20protein%20expression.pdf?la=en Promega protocol]
Expression of Laccase
- Chassis: Promega's E.Coli KRX
- Medium: Autoinduction-Medium supplemented with Chloramphenicol (final concentration 60 μg mL-1)
Cultivation with E. coli KRX in shaking flask(with baffles):
- 200 mL culture in 1000 mL shaking flask with baffles (Schott) with silicon plugs
- Cultivation temperature: 37 °C
- Autoinduction-medium with 20-60 mg L-1 chloramphenicol and if nessassary with 100-300 mg L-1 ampicillin
- Shaking at 140 rpm
- for characterizations: automatic sampling every 30 min
Bioreactor cultivations with E. coli KRX
To obtain higher amounts and concentration of proteins we cultivated and expressed in a bioreactor. It is possible to cultivate several liters and to control temperature, pH and pO_2.
- Bioreactor: Braun Biostat B Bioreactor (3L), Infors Labfors Bioreactor (3L), Bioengineering NLF22 Bioreactor (7 L),
- Autoinduction-medium with 60 mg L-1 chloramphenicol
- Culture volume: 3,0-6,0 L
- Starting OD600: 0.1 - 0.2
- Airflow: 5 NL/min
- pO2-Control: 30 % airsaturation (controlled with stirrer cascade starting with 200 rpm)
- pO2=100% calibration with 300rpm
- pH: 7.0 (controlled with 2M phosphoric acid and 2 M NaOH)
- Antifoam: BASF pluronic PE-8100
- Harvest after 12-13 h
Cell Harvesting
- Harvest cells by centrifugation at 10,000 g for 10 min at 4 °C
- if the purification should start the next day store the cell pellet at 4°C !(the laccase must not be frozen!)
- Resuspend the pellet in 5 mL special buffer or binding buffer for each gramm of cell paste
Solubilization of inclusion bodies
- centrifugation of the celllysate at 40,000 g for 30 minutes
- resuspend the pellet of the lysate in 1 mL 6M Urea solution, incubation for 1 hour
- centrifugation for 10 minutes at 10,000 rpm
- resuspend the pellet in SDS running buffer
Cell disruption strategies
B-PER lysis (chemical lysis)
B-Per bacterial Protein Extraction Reagnt was used for a cell disruption screening according to the following protocol of Thermo Scientific.
- add 4 mL B-Per Reagent per gram of cell pellet
- resuspend the cell pellet by pipetting the suspension up and down until it is homogenous.
- incubate the solution for 10- 15 min at room temperature
- centrifuge lysate at 15.000g for 10 min
- decant the supernatant in a clean tube
The lysate is ready for a following purification step.
enzymatical lysis with lysozym
The lysis with lysozym was used for a cell disruption screening. The following protocol was utilized:
- resuspend the cell pellet in 600 µL of lysozym-solution per gramm
- incubate the solution for 1 h at 4°C
- centrifuge the lysate toseperate soluble proteins from insoluble proteins and cell debris.
combination of chemical and enzymatical lysis
B-Per bacterial Protein Extraction Reagnt was used for a cell disruption screening according to the following protocol of Thermo Scientific.
- add 4 mL B-Per Reagent per gram of cell pellet
- add 2µL of lysozym-solution(50 mg mL -1) and 2 µL DNaseI (2500 U mL -1) per mL B-Per Reagent. (For laccases:Do not use EDTA!)
- resuspend the cell pellet by pipetting the suspension up and down until it is homogenous.
- incubate the solution for 10- 15 min at room temperature
- centrifuge lysate at 15.000g for 10 min
- decant the supernatant in a clean tube
The lysate is ready for a following purification step.
Mechanical lysis
The method of choice to disrupt the cells depends on the amount of biomass.
Mechanical lysis of the (shaking flask) cultivation
Sonication
- Sonication of the re-suspended pellet on ice
- cycle number depends on the volume of the resuspended cells (e.g. 3 mL means 3 cycles)
- one cycle means sonification treatment for 1,5 min with Sonifier 450 by Branson, max. 50 %, cooled on ice, make sure not to heat the cells too much
Precellyse 24 homogenization
- homogenization with the Precellyse 24
- fill the precellyse tubes with a sample volume between 1 mL up to 1,5 mL (for 2 mL tubes)
- homogenize the samples for 3 cycles (6500 rpm for 35 sec. ), to make sure not to heat the cells to much, the sample were stored for 5 min in ice between 2 cycles.
Mechanical lysis of the (bio-reactor) cultivation
Cell disruption with a high-pressure homogenizer
- high-pressure homogenisation with a Rannie Homogenizer:
- disruption of the cells by 3 cycles with cooling phases between the cycles, pressure = 1200 bar, make sure not to heat the cells too much
Purification
Ultra-/Diafiltration

- Arrange the filtration module as shown on the right side.
- Microfiltration (0.22 µm) or cross flow filtration with 300 kDa (we used a Milipore Pellicon XL 300) membrane of sample before ultrafiltration.
- For concentrating the sample just filter it until the desired volume is left in the feed reservoir. For diafiltration (e.g. buffer exchange, desalting) dilute the feed reservoir several times and filter continously.
- Used membranes: [http://www.millipore.com/catalogue/module/C7493 Milipore Pellicon XL 50] or XL 100 membranes
- 50 kDa cut-off
- 50 cm2 filtration area
- tangential flow filter
- Hydrophilic polyvinylidene fluoride membrane
- Used pump: SciLog TANDEM 1081 peristaltic pump
- flow rate during filtration: 40 mL min-1
His-tag affinity chromatography
- For buffers see here
Syringe method
- Column: 1 mL HisTrap FF crude by GE Healthcare
- Equilibrate with binding buffer (10mL)
- Load sample onto column (max. 6 mL)
- Wash with 10 mL binding buffer
- Elute with 5 mL of elution buffer (500 mM imidazole)
- Collect the eluate in 1 mL fractions, the purified protein is most likely in the first or second fraction
- Re-equilibrate the column with binding buffer
ÄKTA method
- Columns:
- 15 mL HisTrap FF crude by GE Healthcare
- 50 mL TALON-Histag-Purification Resin by Clonetech
Column preparation
- If Column is not loaded with Ni-ions /Cobalt-ions:
- Wash column with 5 - 8 Columnvolumes (CV) of deionized water
- Load column with metal-ions(4 CV)
- For HisTrap FF crude: 1,4% NiSO4-Solution
- For TALON-Histag-Purification Resin: CoCl2-Solution
Chromatography protocol for the Äkta-system
- Wash column with 10 CV of deionized water
- Equilibrate column with 10 CV of binding buffer
- Load column with supernatant of the lysed cells (Collect the Flow through for SDS-PAGE analysis)
- Wash Column with 10 CV of binding buffer (Collect the Flow through for SDS-PAGE analysis)
- Elute Protein with an increasing elutionbuffer ratio (gradient 0%-100%, length 200mL)
- Collect the eluate in 10 mL fractions
- Elute remaining proteins with 100% Elutionbuffer (4 CV)
An typical chromatogram of purified laccases is illustrated in the following grafic:
| 55px | | | | | | | | | | |
 "
"