Team:UT-Tokyo/Project/H2 E.coli/System
From 2012.igem.org
H2 E.coli: System


Here we introduce the details of the system we thought.
mixed acid fermentation
As we stated in the background page, our H2 E. coli project aims to produce hydrogen from garbage efficiently using Escherichia coli.
E. coli naturally produce hydrogen through a process called mixed acid fermentation.
We thought we would be able to increase H2 production of E. coli by enhancing a specific pathway of mixed acid fermentation.
Here is an overview of the mixed acid fermentation pathway of E.coli.
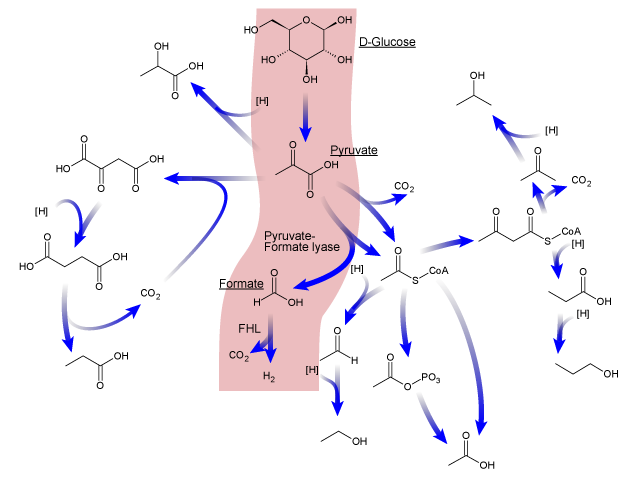
First, Phosphoenolpyruvate (PEP) is produced from glucose by glycolysis, which is in turn transformed into pyruvate or into oxaloacetate. These two molecules are then transformed into several products after a chain of reactions, including hydrogen. ( produced from formate in order to regulate the intracellular pH of E.coli.)[7] So, we expected to be able to increase hydrogen production by overexpressing specific genes of the mixed acid fermentation pathway, that are involved in enhancing H2 production.[2]
Here are the three main reactions which lead to hydrogen production.
- 1:The conversion of phosphoenolpyruvate into pyruvate
Phosphoenolpyruvate + ADP + 2H+ ↔ Pyruvate + ATP
- 2:The conversion of pyruvate into formate
Pyruvate + Coenzyme A ↔ Formate + Acetyl-CoA
- 3:The break-down of formate producing hydrogen
Formate + H+ ↔ CO2 + H2
After we examined a lot of published research, we concluded that the most important reaction of these three reactions in order to increase H2 production is the transformation of formate into hydrogen.
FHL complex and FhlA
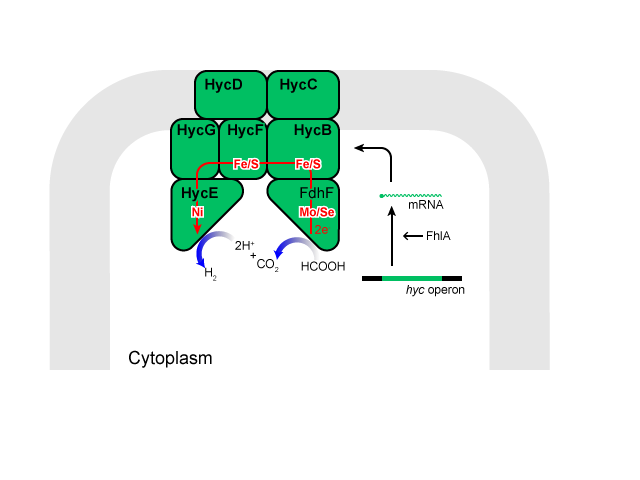
Hydrogen is produced from formate by the formate hydrogenase lyase system (FHL), an intracellular membrane-bound complex, most of the components of which are produced by the hyc operon.
Formate dehydrogenase H[5], encoded by fdhF, converts formate to 2H, 2e, and CO2. Hydrogenase 3 (Hyd-3)[4], encoded by hycE and hycG, is reported to synthesize molecular hydrogen from 2H and 2e. Electron transfer proteins encoded by hycBCDF, are thought to shuttle electrons between formate dehydrogenase H and Hyd-3.
This complex eventually catalyzes the following reaction[6]:
Formate + H+ ↔ H2 + CO2
Here is a simplified model of that complex.
We thought that increasing production of the FHL complex is the best way to boost H2 production. However, doing so by cloning genes of the complex and overexpressing them from a plasmid seemed challenging as the complex consists of a large number of sub-units. Instead, we decided to overexpress a gene fhlA involved in transcriptionally activating genes of the FHL complex.[1]
Additionally, it has been reported that the amino acid replacements E363G in FhlA increased hydrogen production six-fold, compared to native FhlA.[3] So we modified FhlA in the same way in order to further enhance H2 production.
Material and Method
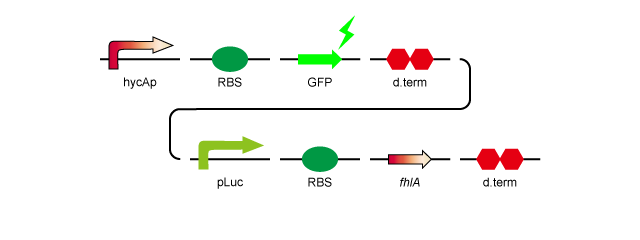
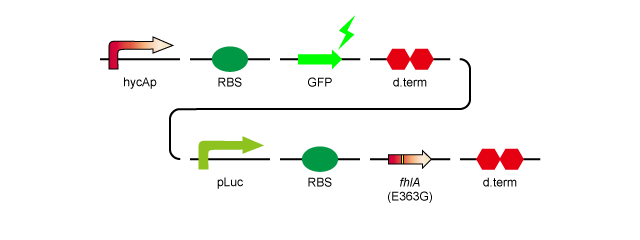
To over-express fhlA, we decided to insert it into plasmid pSB1A2 with a RBS, a lac promoter and a double terminator. We also constructed a device in which hycAp, the natural target of the FhlA protein, promotes GFP expression in order to confirm that overexpression of fhlA results in expression from the hycA promoter, without having to directly quantify the amount of H2 produced.
Here is a model of the construct.
- 1.hycAp-RBS-GFP-d.term-pLac-RBS-fhlA-d.term:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K778010 BBa_K778010]
- 2.hycAp-RBS-GFP-d.term-pLac-RBS-fhlA(E363G)-d.term:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K778011 BBa_K778011]
First, we tried to clone fhlA by PCR using specific primers to the wild-type genome of E. coli.(Jm109) However, unfortunately, the fhlA gene contains an Pst1 restriction site. Therefore, we had to introduce a silent mutation in the sequence of this restriction site by Site-Directed Mutagenesis using inverse PCR. We called this fhlA with a silent mutation mutationfhlA.(mfhlA) Next, we introduced mutagenesis at the codons corresponding to the amino acid replacements E363G in mFhlA by Site-Directed Mutagenesis using inverse PCR. We named this gene mfhlA(E363G). The hycAp sequence was also obtained by PCR using specific primers and the wild-type genome of E.coli. (Jm109)
The RBS, the promoter, the double terminator and the GFP that we selected are the following.
- pLac: BBa_R0011
- RBS: BBa_B0032
- d.Term: BBa_B0014
- RBS-GFP (+LVA Tag)-Terminators: BBa_I763020
After obtaining the genes we wanted by PCR, we ligated them.
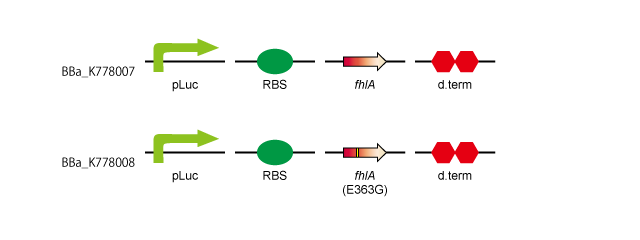
- 1. pLac-RBS-fhlA-d.term:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K778007 BBa_K778007]
- 2. pLac-RBS-fhlA(E363G)-d.term:[http://partsregistry.org/wiki/index.php?title=Part:BBa_K778008 BBa_K778008]
Then, we inserted these into the pSB1A2 plasmid. We cultivated E.coli with the plasmid in anaerobic conditions, because the genes coding the FHL complex are expressed only under anaerobic conditions, and assayed the amount of H2 produced by using Gaschromatography(GC).
We assayed the amount of H2 produced in three types of E.coli: wild type, mfhlA and mfhlA(E363G) .
Results are here.
Full Methods of DNA work are here.
Assay methods are here .
References
- [1] TOSHINARI MAEDA and HIROAKI I. OGAWA -Metabolic Engineering and Protein Engineering for Escherichia coli to Enhance Bacterial Hydrogen Production-Journal of Environmental Biotechnology Vol. 9, No. 2, 69–74, 2009
- [2] Toshinari Maeda & Viviana Sanchez-Torres & Thomas K. Wood -Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli-Appl Microbiol Biotechnol (2007) 77:879–890 – September 2007.
- [3] Viviana Sanchez-Torres,1 Toshinari Maeda,and Thomas K. Wood -Protein Engineering of the Transcriptional Activator FhlA To Enhance Hydrogen Production in Escherichia coli- APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Sept. 2009, p. 5639–5646
- [4] Bagramyan K, Trchounian A (2003) Structural and functional features of formate hydrogen lyase, an enzyme of mixed-acid fermentation from Escherichia coli. Biochemistry (Mosc) 68:1159–1170
- [5] Axley MJ, Grahame DA, Stadtman TC (1990) Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem 265:18213–18218
- [6] Woods DD (1936) Hydrogenlyases: the synthesis of formic acid by bacteria. Biochem J 30:515–527
- [7] Böck A, Sawers G (1996) Cellular and Molecular Biology. In: Neidhardt FC, Curtiss JR II, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (eds) Escherichia coli and Salmonella. 2nd edn. ASM Press, Washington, pp 262–282
 "
"