Team:Cambridge/Lab book/Week 3
From 2012.igem.org
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|
Contents |
Monday (09/07/12)
- Bacillus salts *10 made up and autoclaved.
- Medium A base *10 made up (glucose will be added tomorrow)
- bacillus strain 168 streaked out and grown on plate overnight.
Tuesday (10/07/12)
- Filtered glucose added to sterile Medium A base.
- Sterile aliquots of salts and medium apportioned and put in fridge (10*50ml each)
- Needed to make up MgCl2 and CaCl2 solutions of the correct concentrations. Required multiple dilution steps:
- MgCl2 (250nM), Mr = 147 Da, make 100ml of solution
- 0.508g of MgCl2 dissolved in 100ml of H2O
- 1ml of this solution mixed with 99ml of H2O
- 0.1ml of this solution mixed with 99.9ml of H2O
- CaCl2 (50mM), Mr = 203.3 Da, make 100ml of solution
- 0.735g of CaCl2 dissolved in 100ml of H2O
- Made up Medium A (150ml) and Medium B (50ml)
Wednesday (11/07/12)
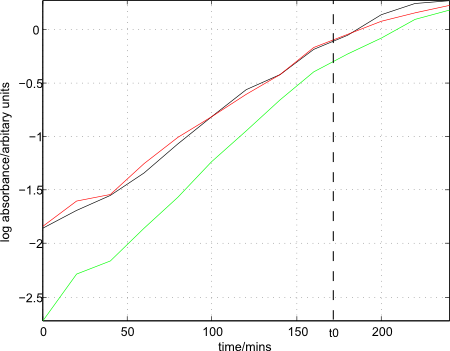
- Colonies of strain 168 removed from plate and added to three separate conical flasks, each with 48ml of Medium A inside. OD650 readings taken until enough cells were added such that absorbance was between 0.1 and 0.2.
- Flasks placed inside a shaking incubator (200rpm) and samples taken every 20mins until growth had leveled off, as determined by the concurrently plotted growth curves.
- Growth curves, as well as t0 (at 170mins) plotted:
- 90 mins after t0 (at 260 mins), cells removed from shaking incubator.
- 0.45ml of Medium B pre-warmed in Eppendorf tubes.
- 20 individual samples of 0.05ml taken from each growth flask and added to Eppendorfs.
- Eppendorfs placed back in shaking incubator for 60 mins with lids off.
- Eppendorfs centrifuged at ~13000 rpm for 10 mins, supernatant removed.
- 60% glycerol added (0.5ml/tube) and tubes vortexed.
- All 60 tubes now frozen at -80 °C.
- Edited protocol so that it actually works.
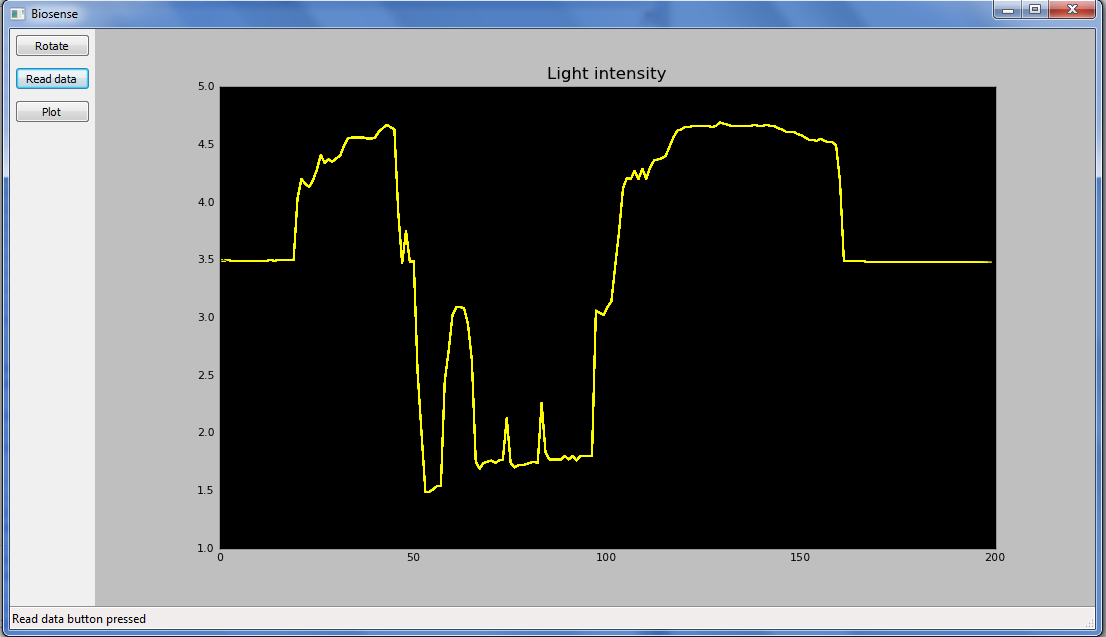
Arduino circuitry
- Real-Time image captured by our freshly made primitive software, monitoring a light sensor on an arduino board. C++ was used to communicate with the Arduino and Python was used for data logging.
Thursday (12/07/12)
Test transformation of frozen bacillus stocks
- Two samples of bacillus defrosted from different batches.
- Supernatant spun off, bacterial pellet isolated and 0.1ml of Medium B added.
- Plasmid Tag RFP-T unfrozen. Concentration of DNA = 360ng/μl, so 2μl needed for the desired 0.6μg of DNA. DNA added to bacteria.
- Eppendorfs placed in shaking incubator at 30 °C and 180rpm for 60 mins.
- Bacteria plated on choramphenicol containing plates. Aseptic technique was used as far as possible.
Friday (13/07/12)
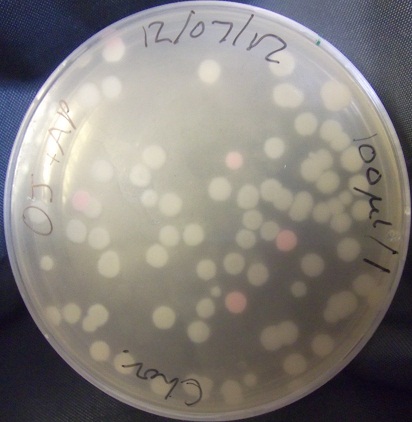
Test transformation of bacillus stocks
- Results: In this image, several colonies have clearly gained pink coloration from the transfected plasmid. This demonstrates that our stocks should be usable. However, the large number of opportunistic colonies that do not appear to have been transfected means that we will have to be careful to check that our transfected bacteria contain what is expected.
Transformation of bacillus and e.coli with lux genes from 2010
- Progress so far: (1) made 25ug/ml chloramphenicol plates; (2) Transformed E. coli and B. subtilis using the corresponding Protocols with the Vibrio LuxBrick from Cambridge 2010 ([http://partsregistry.org/Part:BBa_K325909 BBa_K325909]); (3) Plated the transformed cells and incubate at 37 degrees celsius overnight
- Nanodrop was used to determine the concentration of DNA in the biobrick after resuspension. The result reading was 103ng/ul.
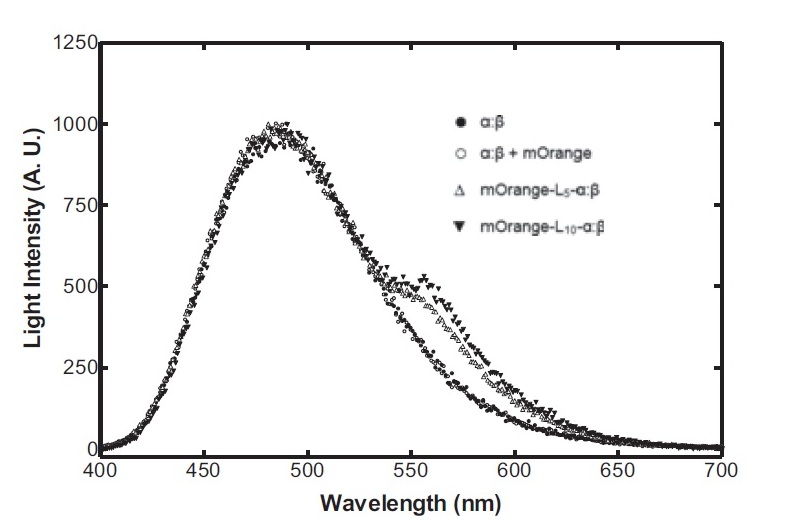
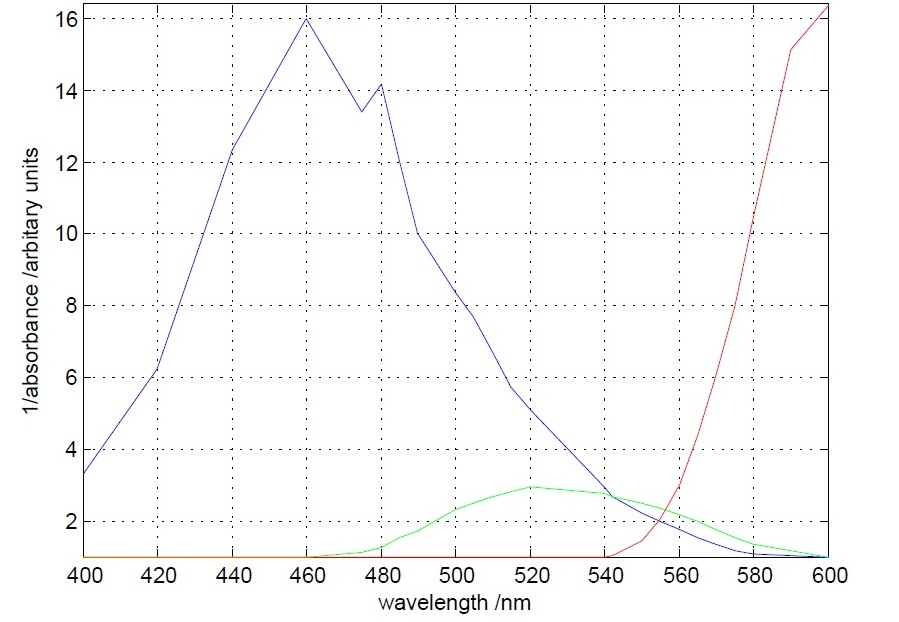
Testing of the spectral properties of filters
- Blue , Green and Red filters were tested in different wavelengths (400 nm - 600 nm) in a spectrophotometer.
- Excellent absorbance/emmitance results given by the blue and red filters near 580nm and 490nm respectively. 580nm is the wavelength where intensity of light is to be measured to identify the presence of mOrange. 490nm is the wavelength where maximum light intensity is expected with or without the presence of mOrange.
 "
"