Team:Cambridge/Lab book/Week 4
From 2012.igem.org
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|
Contents |
Monday (16/07/12)
Ratiometrica- Lux: Transformation of bacillus and e.coli with lux genes from 2010
- Arabinose induction: from Cambridge 2010 iGEM Team's data, the arabinose concentration which will give the optimum amount of light is around 3mM. We hence added 0.45mg arabinose to each mL of LB.
- Light is detected after around 5 hours in the transformed E. coli
- B. subtilis plates have no growth, very possibly due to plasmid backbone pSB1C3 not being compatible in B. subtilis
Tuesday (17/07/12)
Ribosense: PCR of vector
- Plasmid PJS 130 used as our backbone for isolating the magnesium riboswitch.
- Primers:
- Forward: TTCAAAACATGACCTATGACgtcgcagagtatgccg
- Reverse: cctccctctgctaaaacACAAGGCATTAACACTACAT
- PCR settings:
- 95 °C - 6mins
- 98 °C - 10secs
- 58 °C - 45secs
- 72 °C - 180secs
- Repeat above 35x
- 72 °C - 5mins
- 25 °C - 1min
Ribosense: PCR of riboswitch DNA from bacillus genome
- Colony of strain 168 used as template for this experiment.
- Primers used:
- Forward: cggagggagacgattttgTGTTCCGTAATTGTGATGTAAG
- Reverse: acaccggcatactctgcgacGTCATAGGTCATGTTTTGAACC
- PCR settings - as above (run in parallel).
Wednesday (18/07/12)
Ribosense: Gel electrophoresis of PCR products
- PCR fragments for Mg2+ riboswitch from yesterday separated on gel. 90mins at 100V.
- Lane 1: Ladder
- Lane 2 + 3: Riboswitch DNA from genome (replicates) - 550bp fragment expected.
- Lane 4 + 5: Vector DNA from pJS130 (replicates)- Chloramphenicol resistance marker - 9.0kbp fragment expected
- Lane 6: Postitive control - 1.9kbp fragment expected
- Riboswitch DNA was successfully amplified. However, vector DNA was not. Given that the postive control was also successful, it seems likely that something went wrong with some stage before the PCR amplification. We will try to rectify the problem tomorrow.
Ribosense: Purification of riboswitch DNA from gel
- Successful riboswitch DNA extracted from gel and purified using a minelute column. DNA frozen for later Gibson assembly.
Ratiometrica-Lux: Transformation of e.coli with lux genes from 2010
- Progress: (1) Streaked 8 25ug/ml Chloramphenicol plates with transformed e.coli and incubate overnight; (2) inoculate individual colonies from the plates in liquid medium; (3) Incubate overnight at 37 degrees
Thursday (19/07/12)
Ribosense: PCR of vector
- Plasmid PJS 130 used as our backbone for isolating the magnesium riboswitch.
- Primers:
- Forward: TTCAAAACATGACCTATGACgtcgcagagtatgccg
- Reverse: cctccctctgctaaaacACAAGGCATTAACACTACAT
- PCR settings:
- 95 °C - 6mins
- 98 °C - 10secs
- 55 °C - 45secs
- 72 °C - 240secs
- Repeat above 35x
- 72 °C - 5mins
- 25 °C - 1min
- Settings changed - Annealing temperature and elongation time - attempting to debug PCR.
Ribosense: Gel electrophoresis of PCR products
- PCR fragments for Mg2+ riboswitch from yesterday separated on gel. 90mins at 100V.
- Lane 1: Ladder
- Lane 2, 3 + 4: Vector DNA from pJS130 (replicates) - 9.0kbp fragment expected
- Lane 5: Postitive control - 1.9kbp fragment expected
- Lane 6: Negative control - no fragments expected
- Once more, the PCR appears to have failed. We will try to redesign the primers tomorrow to make them functional.
Friday (20/07/12)
Ratiometrica-Lux: Transformation of e.coli with lux genes from 2010
- Arabinose induction: Using the same 3mM arabinose prepared earlier in the week, the E. coli were spun down and the arabinose was added. After 5 hours, light is observed.
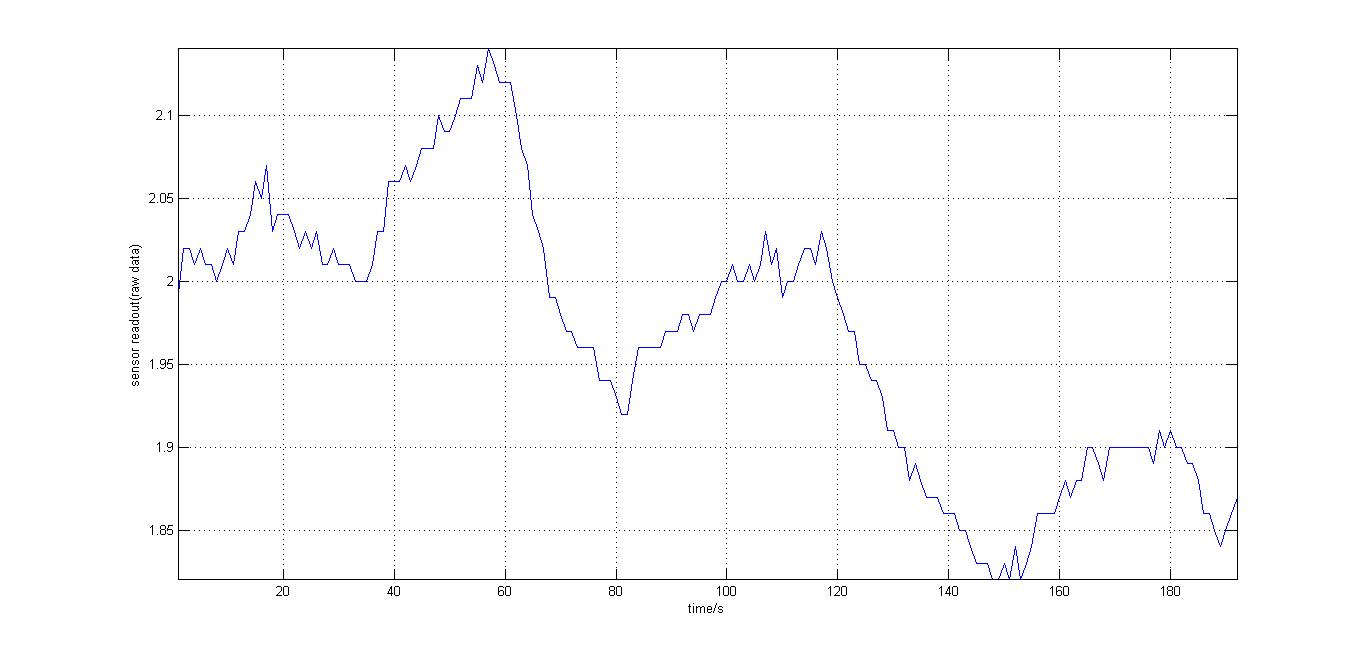
- Testing the Arduino kit: the testing took place in the dark room, and the cell culture (in 1.5mL Eppendorf tubes) was held close to the photosensor and then taken away repeatedly. The data recorded was plotted with Matlab, as shown:
- It should be noted that the background light levels were falling linearly, possibly due to negative feedback on the photosensor, but there is a consistent 5-6% increase of the original base reading upon placing the cells close the the photosensor.
 "
"