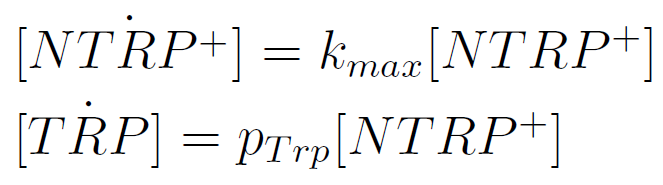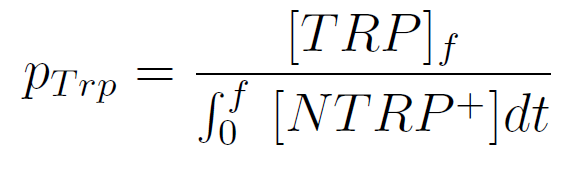Team:Buenos Aires/Results/DevicesTesting
From 2012.igem.org

Contents |
Devices testing
Experimental Setup
Plasmids and BBs
In order to construct the yeast expression plasmids we choosed 3 vectors, 2 with a tryptophan marker and 1 with an histidine marker:
- pCM182/5, which are centromeric plasmids with TRP1 marker, and with a doxycycline repressible promoter [Gari et al 1996].
- pEG202, with a 2 ori, HIS3 marker and a constitutive promoter (PADH1).
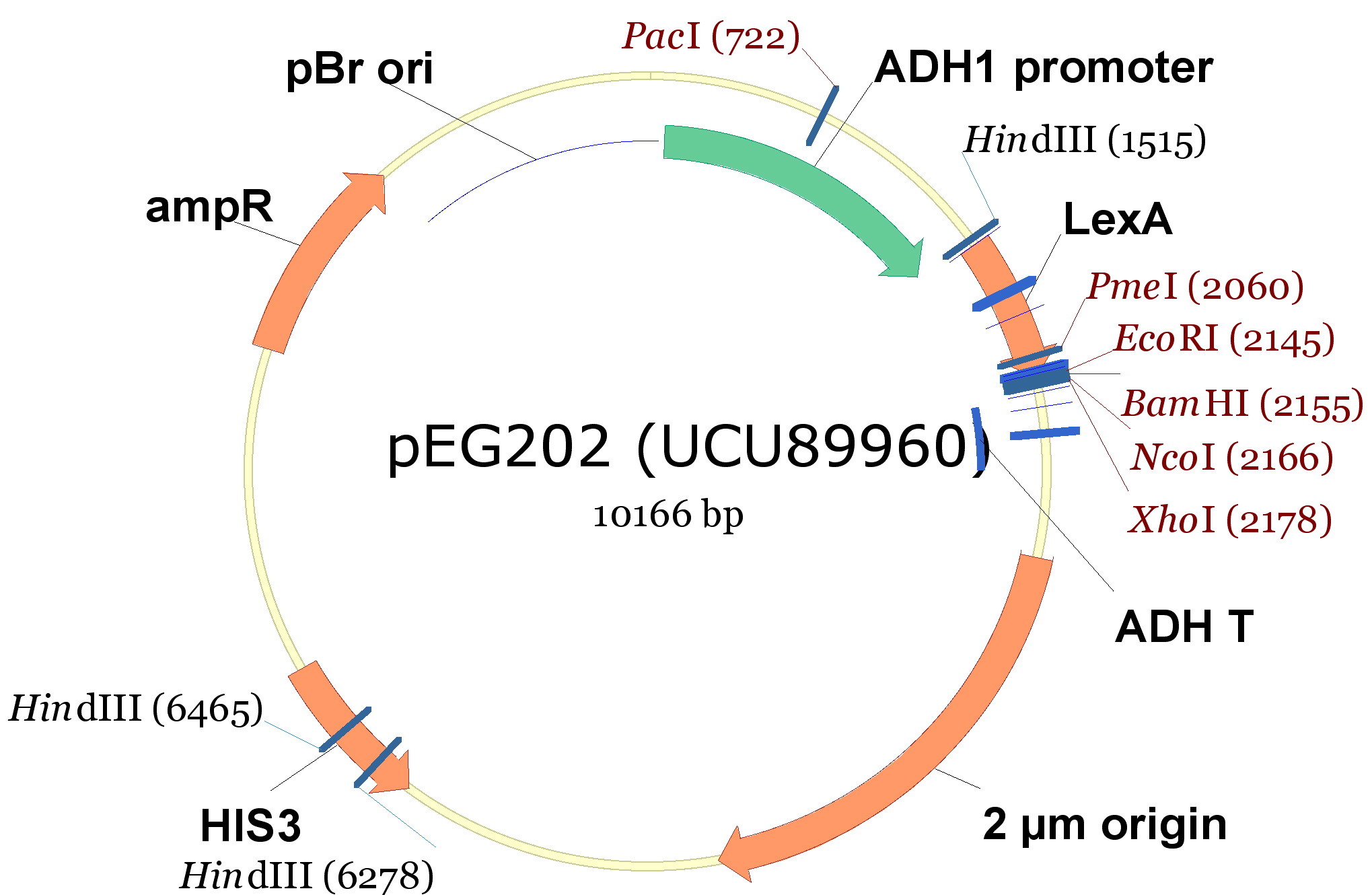
| 340px |
The cloning we did was:
| <partinfo>BBa_K792009</partinfo> (PoliHa) | <partinfo>BBa_K792010</partinfo> (TRPZipper2) | <partinfo>BBa_K792011</partinfo> (PoliHb) | <partinfo>BBa_K792012</partinfo> (PoliWb) | |
|---|---|---|---|---|
| pCM182 (TRPa) | X | X | ||
| pCM185 (TRPb) | X | X | ||
| pEG202 (HIS) | X | X |
Cloning protocol
- Digestion of plasmids and TRP/HIS export device:
- pCM182; pCM 185; BBa_K792010 y BBa_K792012 were digested with BamHI and Pst1 restriction enzymes.
- pEG202; BBa_K792009 y BBa_K792011 were digested with BamHI and Not1.
- Purification of digested vectors (pCM185; pCM182; pEG202)
- Ligation of vectors and devices according the anterior table (T4 ligase protocol, overnight)
- Transformation of E. Coli DH5a with the ligation products. Bacterias were plated on LB-agar with Ampicillin, and incubated over night at 37 °C.
- Colonies were used for liquid cultures (LB + Ampicillin) and minipreps were made.
- Constructions (vector + insert) were checked by digestion with restriction enzymes, and 1%-agarose gel (1kb and 100bp as markers).
Yeast strains
Once obtained the desired constructions, we transformed yeast strains:
- TCY3081: W303, bar1-, ura3::PAct1-YFP
- TCY3128: W303, bar1-, leu2:: Pprm1-CFP 405
Final transformed strains
We got the following transformed strains:
|
|
|
|
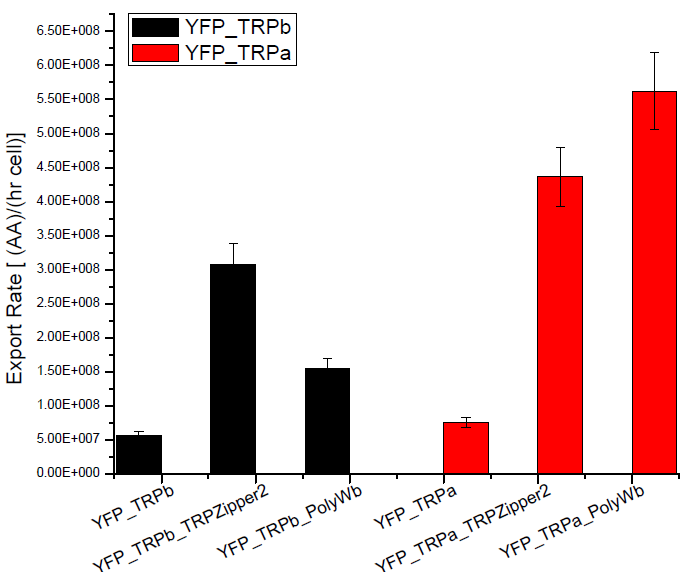
Secretion Rate of Trp as a function of culture growth
The first step was to actually check if the construct works: do the transformed yeast strains - with the tryptophan devices- actually secrete tryptophan into the medium?
To test this we used the following strains:
|
| ||||||||
|
Protocol
- We started 5ml cultures with 3 replica until they reached exponential phase, overnight, using a -T medium.
- Starting OD for the assay 0.1 (exponential phase).
- We measured OD every hour until they reached an OD: 0.8 (5 hs approximately).
- We measured the Trp signal for each culture medium using the spectrofluorometer.
NOTE: The fluorescence meausurements taken for the Tryptophan in the medium, take into account both the aminoacid secreted by the device and the one diffused.
Therefore the secretion rates calculated will be higher than the actual ones. We used an empty plasmid as control to study tryptophan diffusion.
We used a simple model to measure the secretation rate for all the strains. Since the cultures are in exponential phase, we take
After a few calculations, we find that
Results
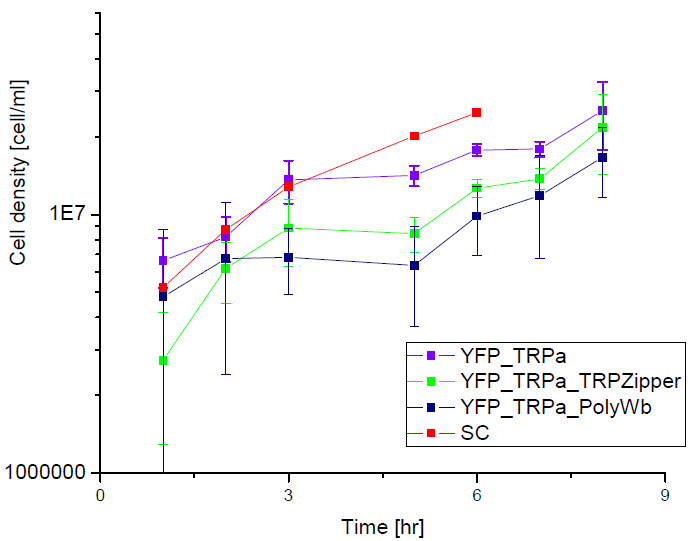
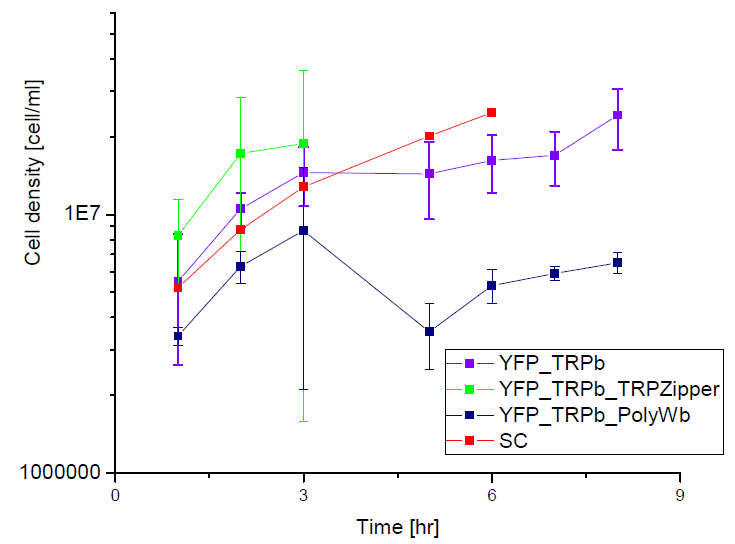
Next we show the average OD for each strains used, needed to calculate the export rates of Trytophan.
Figure1. check
Figure2. check
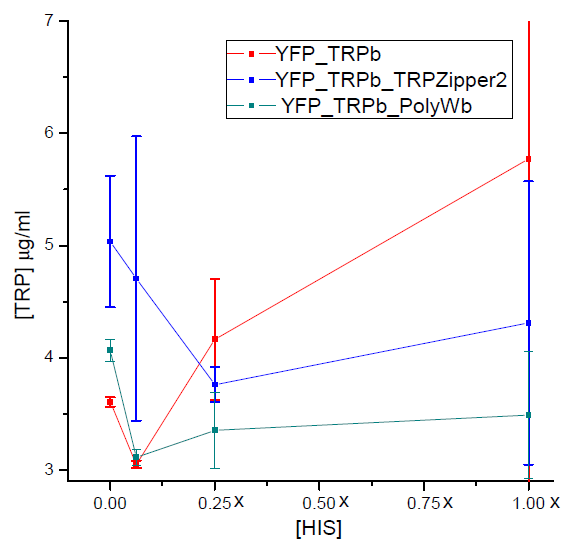
Tryptophan secretion at increasing histidine concentrations
We asked ourselves which was the dependance of tryptophan secretion on the amount of histidine in medium. We carried on this test in order to determine whether the secretion of tryptophan depends on the concentration of another aminoacid in the media, such as histidine and what would be the necessary amount of histidine in medium for the start of our system.
In this experiment we used strains:
| YFP_TRPb_TRPZipper2 | YFP_TRPb_PolyWb | YFP_TRPb (control) |
Protocol
- Starters of each strain used were grown over night in -T media, at 30 °C in shaker.
- After 12 hs, cells were pelleted and washed with -HT media.
- Cultures with increasing concentrations of histidine (0X, 1X, 1/4X and 1/16X) were set at an initial OD: 0.1, for each strain with 2 replica.
- We left the cultures in shaker at 30ºC for 5 hours. After that time, we measured the final OD reached by cultures with the use of a spectrophotometer and the amount of tryptophan present in medium with a spectrofluorometer.
Results
Figure 3. Final tryptophan concentration as a function of histidine concentration in the media.
With this result we can infer that tryptophan export has no clear dependence on histidine concentration in the media. Therefore, supporting the assumption of the model.
Measurement of Trp in medium and Basal Production
To check the efectiveness of our biobricks, we must first determine the ammount of tryptophan secreted by natural strains to the medium, so we can compare. With that end in mind, we designed a protocol for measurement of tryptophan in medium, based in its fluorescense at 350nm, when excited with 295nm light. As a previous step, we checked that none of the other aminoacids used in the medium interferes, by graphically comparing the spectres for uncomplemented medium and medium complemented with leucine, uracile and histidine, at an appropiate range.
To determine Trp concentration, we must first have a way to transform our readings (intensity) to a more useful output, so we made a calibration curve, through serialized 1:2 dilutions of our medium, which Trp's concentration is 50mg/mL, until approximately constant intensity.
The procedure to measure secretion rates will be growing the strain from a known OD in exponential growth phase in -T medium and plotting it's OD over time, spin-drying at time=t, retrieving the supernatant's Trp concentration and dividing it by the integral of OD vs. time between time=0 and time=t, so we get to a rate which will be proportional to the number of cells in the culture, which means we can actually compare between different strains. Since our medium is free from Trp, all of it should come from within the cells, and if the culture is growing at exponential rates, lysis should be negligible, so the only explanation would be cells exporting their own Trp.
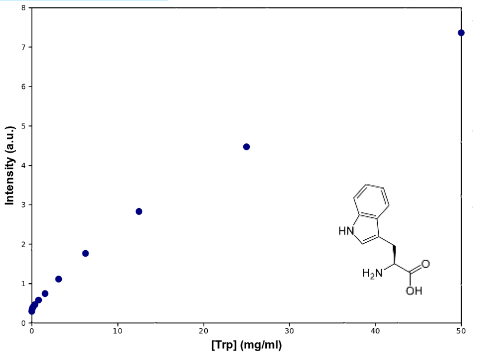
|
| Graph:Tryptophan calibration curve |
Results
As can be seen from the graph the screening of the concentration of the Trp in medium describes an almost lineal function. Through this experiment we can be sure that we would be able to measure increase of Trp in medium as it is exported from the cells, within the biological range of export. The sensitivity of this method seems to be enough to detect concentrations as low as ~0.02mg/mL, and as high as 50mg/mL, maybe more. Since our medium is 50mg/mL, we assume that's the saturation point of the curve. If we get bigger intensities than the one corresponding to it, we will dilute the sample.
Because of time constraints, we haven't been able to check the method with either our designed strains nor the non-exporting ones.
 "
"