From 2012.igem.org
July 2012
S M T W T F S
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
August 2012
S M T W T F S
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
September 2012
S M T W T F S
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
October 2012
S M T W T F S
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
June 07
Transformation (LSE)
Transformed DNA:
lacZ (well 4:12G, I732019)
p + lacO (well 1:6G, R0011)
Cells: neb10beta (donated)
Protocol from: http://www.neb.com/nebecomm/products/productc3019.asp
Controls: puc19, no DNA (8 plates)
June 08
Transformation results
puc19: growth
negative control: no growth
lacZ, lacO: possible small colonies
liquid culture in amp media (100 ug / ml):
no growth of lacZ, lacO
growth of puc19
June 12
DH5a Competent Cell Prep
Streak plated cells on LB no amp plate, let grow overnight
June 13
Transformation (LSE)
Transformed DNA:
lacZ (well 4:12G, I732019)
p + lacO (well 1:6G, R0011)
Cells: DH5 alpha (donated)
Protocol from: http://openwetware.org/wiki/Haynes:Assembly101 (30 minute transformation)
Controls: puc19, no DNA (8 plates)
Transformation (istb4, Abhi)
Transformed DNA:
Cells:
Protocol from:
Controls:
DH5a Chemically Competent cell prep
Grew 2 seed colonies from streak plate in LB no amp
Grew controls to test for contamination
Both Seed colonies grew, no contamination present
June 14
Competent cell prep
Prepared CaCl2 buffer solution and CaCl2 glycerol buffer solution
Grew seed colony in 400mL LB no amp
June 15
Competent cell prep
Centrifuged falcon test tubes containing liquid colonies
Resuspended in CaCl2 buffer solution and incubated for 15 mins
Centrifuged and resuspended in CaCl2 glycerol buffer solution
Chilled overnight
June 16
Competent cell prep
Aliquotted 200uL into test tubes
Stored in -80C
June 17
Streak plated prepared competent cells on LB no amp plate
June 19
Transformation (LSE)
Transformed DNA:
T7 promoter BBa_I712074
Constitutive promoter BBa_J23102
Cells: DH5 alpha (donated)
Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
Controls: puc19, no DNA
Plated 2 copies of each (100 ul, 250 ul) on LB amp plates.
Made 50 LB Amp plates.
June 20
Plated negative control on LB Amp plate
Liquid cultures of T7 promoter and constitutive promoter
Transformation (LSE)
Transformed DNA:
RBS (well 1:1H BBa_B0030)
TetR GFP (well 2:8A Part:BBa_I13522)
Cells: DH5 alpha (donated)
Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
Controls: puc19, no DNA
June 21
Made Liquid Cultures of E.coli transformed with RBS B0030
Made Liquid Cultures of E.coli transformed with TetR GFP
miniprepped and nanodropped T7 promoter BBa_I712074 and Constitutive promoter BBa_J23102 liquid cultures
liquid cultures:
RBS1
RBS2 (duplicate_
GFP1
puc19
negative controls
5 ml LB amp
overnight cultures
replated GFP1 & 2 (duplicates)
Nanodropped plasmid DNA samples
Constitutive promoter 1: __ng/uL
Constitutive promoter 2: __ng/uL
T7 promoter 1: __ng/uL
T7 promoter 2: __ng/uL
June 22
Miniprepped liquid cultures: RBS (well 1:1H BBa_B0030) and TetR GFP (well 2:8A Part:BBa_I13522)
Picked colonies:
1 colony from double terminator (dt1) plate
1 colony from t7 polymerase (pol1) plate
1 colony from puc19 plate (positive control)
1 colony from dh5a plate (negative control)
started liquid cultures of each colony (5 mL LB amp each)
June 26
Transformation:
Transformed DNA:
double terminator (B0017, 2:6K)
T7 RNA polymerase (I715038, 2:15C)
puc19, negative control
Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
Cells: dh5a
June 27
6-26 transformation results:
Controls correct
2x terminator: ~19 colonies
RNA pol: 1 colony
Liquid cultures including controls
June 28
Miniprepped double terminator (B0017, 2:6K) and T7 RNA polymerase (I715038, 2:15C) liquid cultures
July 2
Cleaned up liquid waste
Made SOB media
Finalized oligos for magainin construct
July 3
Autoclaved SOB media
Added glucose to make SOC media
Nanodropped double terminator (B0017, 2:6K) [DT1: 24.5, DT2: 29.6] and T7 RNA polymerase (I715038, 2:15C) [P1: 64.6, P2: 55.3] liquid cultures
July 24
Plasmids arrived courtesy of University of Pennsylvania School of Medicine
pET29a vectors containing coding sequence for Topoisomerase mutants CSCS and CSCS D168A described here
July 25
Tranformed CSCS topo 0 plasmid and CSCS D168A topo into DH5a Turbo cells (with neg control and Puc19 neg control)
Plated on Kanamycin plates
July 26
Picked colonies colonies from Topo 0 and Topo D168A and grew liquid cultures in Kan media
July 27
Miniprepped liquid colonies and nanodropped.
Plasmid concentrations
Topo O:
Topo D168A1:
Topo D168A2:
July 30
Prepared Kan Media and Kan Plates
July 31
PCR amplified polylinker sequence of Topo plasmid with Promega GoTaq protocol
Used pET29a upstream forward primer and T7 terminator reverse primer
August 3
Submitted pET29a Topoisomerase plasmid to Biodesign for sequencing
Resuspended GFPT1 and GFPT2 oligos with molecular grade (nuclease-free) H2O.
Final Concentration 100uM
(gfpt1 top1, gfpt2 top1, gfpt1 top2, gftp2 top2, gfpt1 bot1, gfpt2 bot1, gfpt1 bot2, gfpt2 bot2)
(3uL of each oligo + 2uL 10x annealing buffer, 6uL molecular grade H2O. 20uL Reactions)
Heated for 5 minutes at 100C. Let cool to room temperature on the heating block, stored at -20C.
August 10
PCR of Alpha-4, 1-omega, omega, and alpha fragments using corrected primers
August 13
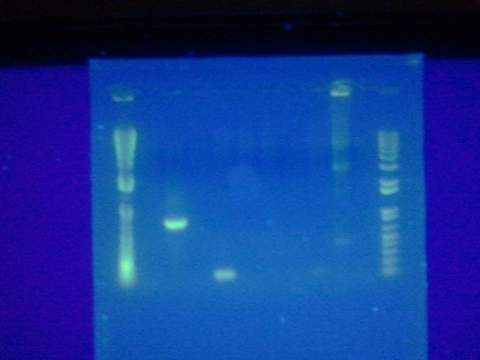
Ran gel of split beta gal fragments. Confirmed 3 out of the 4 fragments except for the alpha-4 fragment.
August 14
Assembled magainin insert via Overlapping oligo assembly
Digested pUC 19 plasmid with EcoRI and PstI
Transformed Magainin insert into digested pUC 19 plasmid. Failed. Probably too much X-gal on plate.
Ran gel for the beta gal alpha-4 fragment. Failed. Fragment not in the correct size-band.
August 15
[http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/topo0_T7Term.seq Topo 0], [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo1_T7Term.seq D168A Topo1], and [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo2_T7Term.seq D168A Topo2] sequence results
August 16
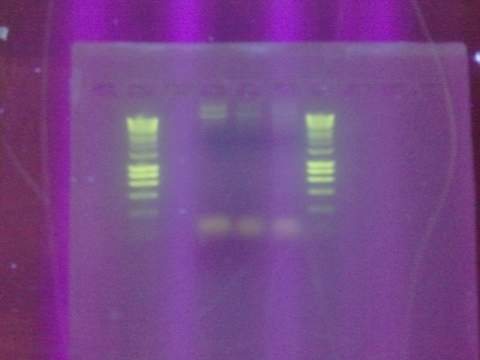
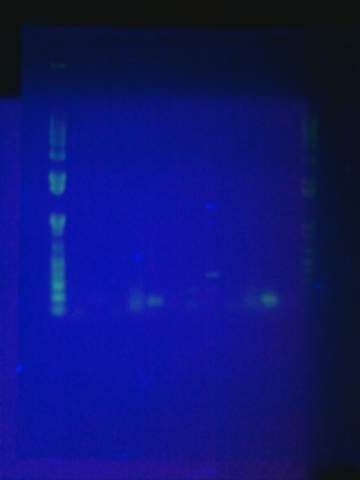
Replated magainin insert + plasmid into the grid. Failed. All blue colonies meaning that no insert. Tried gel for all gel-isolated fragments. Failed. Did not get a band in the 2000 bp region. Only got things below 200 bp.
October 1
Minipreps of:
J61011 + S + L + ALPHA 2B
J61100 + S + L + ALPHA 2B
J61100 + S + L + ALPHA 2C
PLUX2 + RBS + ALPHA 1A
PLUX2 + RBS + ALPHA 1A (2)
PLUX + S + L + ALPHA 2C
J61101 + S + L + ALPHA 1A
J61101 + S + L + ALPHA 1A (2)
PLUX2 + RBS + S + L + ALPHA 2B
PLUX2 + RBS + S + L + ALPHA 2B (2)
J61101 + S + L + ALPHA 2C
PLUX + S + L + ALPHA 2B
Ran Twice when finding concentrations:
J61101 + S + L + ALPHA 1A
J61101 + S + L + ALPHA 2B
Restricted all the above and:
Strep + Linker + OMEGA
Strep + Linker-4 + OMEGA
with:
Ligated into pSB1C3 shipping vector and transformed into BL21(DE3) cells.
Prepared all 14 above samples for sequencing.
 "
"