Team:Cambridge/Lab book/Week 13
From 2012.igem.org
| Week: | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 |
|---|
Contents |
Monday (17/09/12)
Ratiometrica - Lux
Testing of the 2.0 construct:
Numbers 33,35,36 have correct sequence as of being sent to us. However they were not visibly luminescent. *34 was.
Overnight cultures of each of the 6 culture lines sent were set up. Additionally, inducing plates were streaked, including the 2010 lux biobrick as a luminescence control and pjs130 as an iptg induction control.
Primers ordered to ligate 2.0 construct into pjs130 for insertion into bacillus.
Tuesday (18/09/12)
Ratiometrica - Lux
None of restreaked plates visibly luminescent, aside from *34 (faint), even under photon counting camera. Pjs130 not green, so iptg induction potentially didn't work. LuxBB induced and luminescent.
Cultures induced with 1mM iptg or 3mM arabinose for luxBB, imaged after 3 hrs. Some extremely faint luminescence for 33,34,38 on photon counting camera.
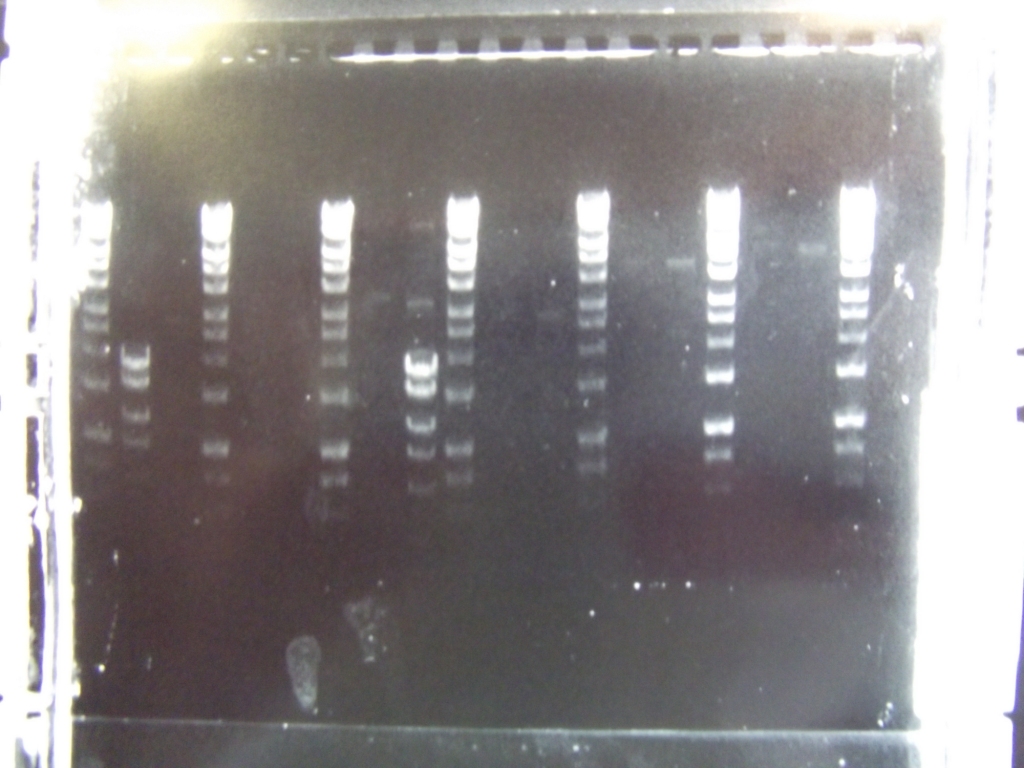
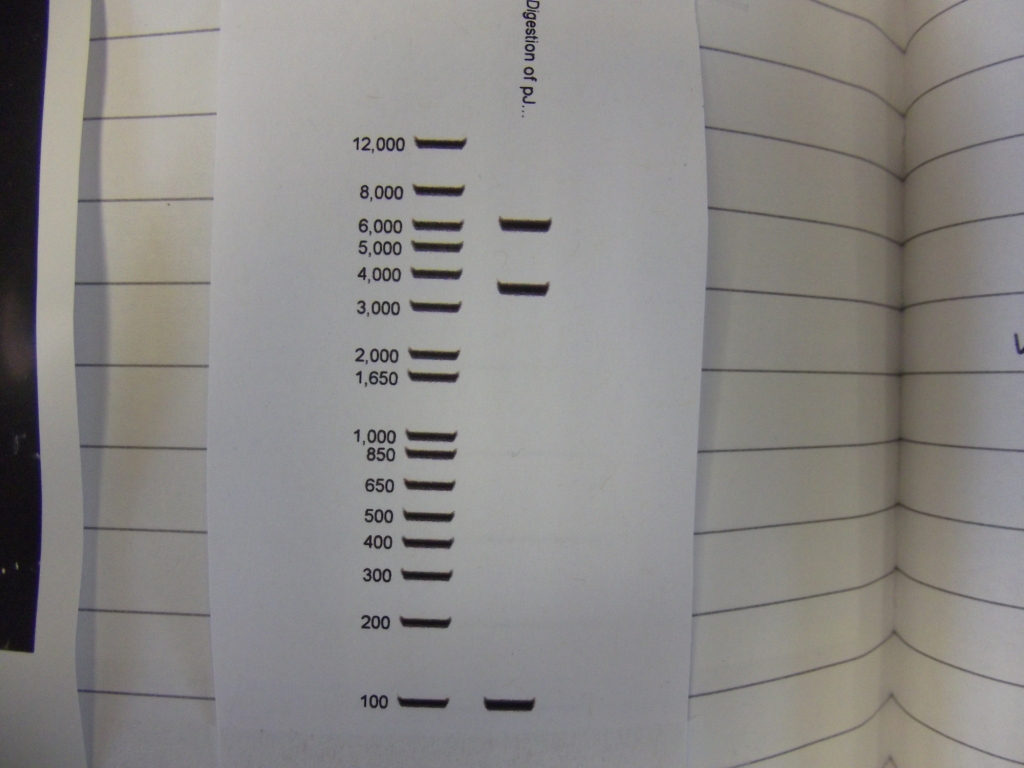
Cultures miniprepped and restriction digests carried out w/spa1. Results are shown. Results indicate problem with the miniprep and/or damage to the construct - expected bands at 6625, 2838, 1876 and 53 bp.
Wednesday (19/09/12)
Ratiometrica - Lux
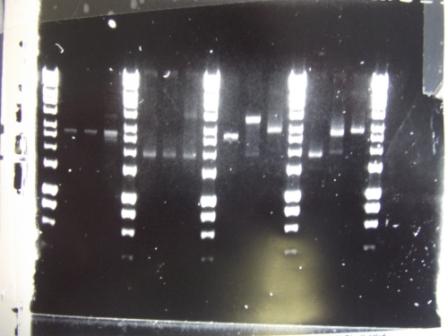
Digest of 2.0 miniprep with different enzyme, hindIII, carried out. 15ul template DNA used in a 20ul reaction. Again, results were odd. Cut and uncut minipreps ran.Cultures of ratiometrica (fluorescent) were miniprepped to be transformed into bacillus. A restriction digest was performed on these minipreps, and produced bands of the correct size.
Thursday (20/09/12)
Ratiometrica - Lux
Transformation of fluorescent ratiometric construct into bacillus was unsuccessful
Transformations of XL1-blue cells with resuspended construct plasmid from DNA 2.0 yielded orange colonies, with some white, and some showing sectoring, indicating insert loss.
Plates all appear to be luminescent. A random selection of colonies was picked and patched in order to investigate whether luminescence cosegregated with orange colour.
Friday (21/09/12)
Initial pick and patch experiments showed cosegregation, but we did not have immediate access to a photon-counting camera or a DSLR capable of long exposures, so we did not obtain images. Representative colonies were streaked out onto a fresh plate to be imaged later.
Saturday (22/09/12)
A PCR was run to amplify several fragments, including:
- The ratiometric luciferase construct, to insert into;
- pSB3C5, a low copy number vector
- Oli's Mg riboswitch construct
All fragments were obtained.
Sunday (23/09/12)
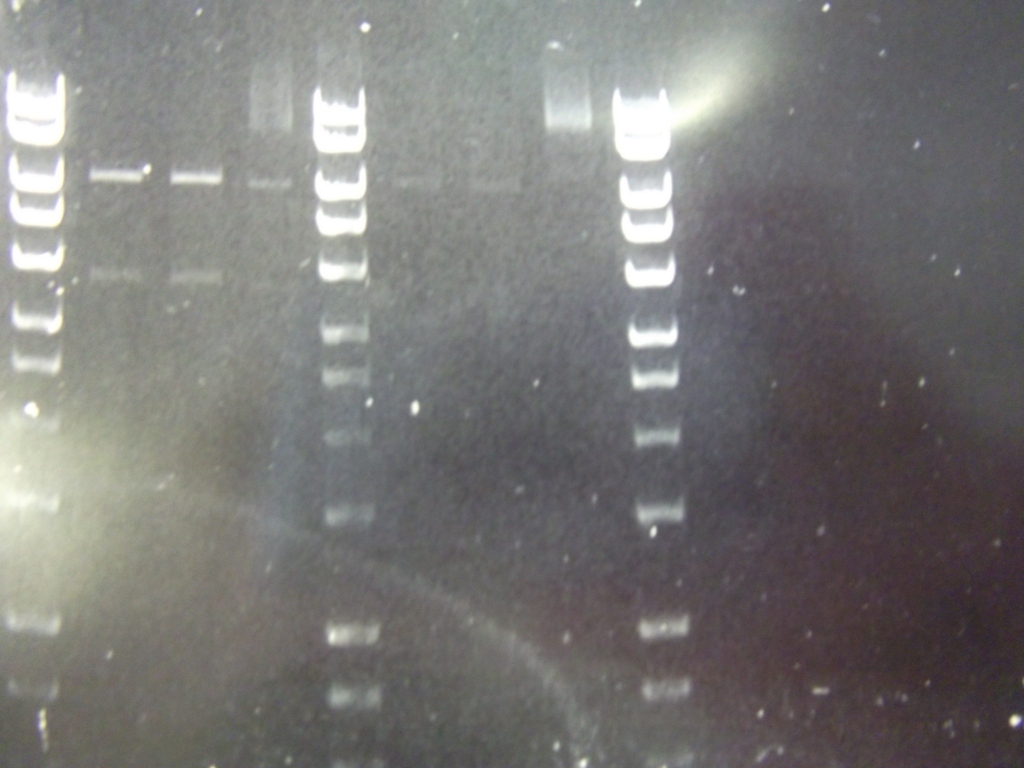
Fragments for the fluoride riboswitch, the spoVA swap biobrick and the insertion of the 2.0 construct into pSB3C5 were all digested for two hours in 50 ul reactions:
- 5ul NEB2
- 1 ul EcoRI
- 1 ul PstI
- 0.5 ul BSA
- 10 ul DNA
- 32.5 ul HPLC H20
Enzyme was heat-inactivated for 20 minutes at 65 C
 "
"