Team:TU Darmstadt/Protocols/GC-MS/Results
From 2012.igem.org
Contents |
Sample preparation
We used two methods for our TPA analytics. Therefore we used standard sample preparation protocol:
Purge and Trap
- OP-1 add. 20 µl in 10 ml Water
- OP-2 add. 20 µl in 10 ml Water
- OP-K add. 20 µl in 10 ml Water
TPA short
- OP-1 add. 1ml in vial
- OP-2 add. 1ml in vial
- OP-K add. 1ml in vial
Results
=TPA Short ATS
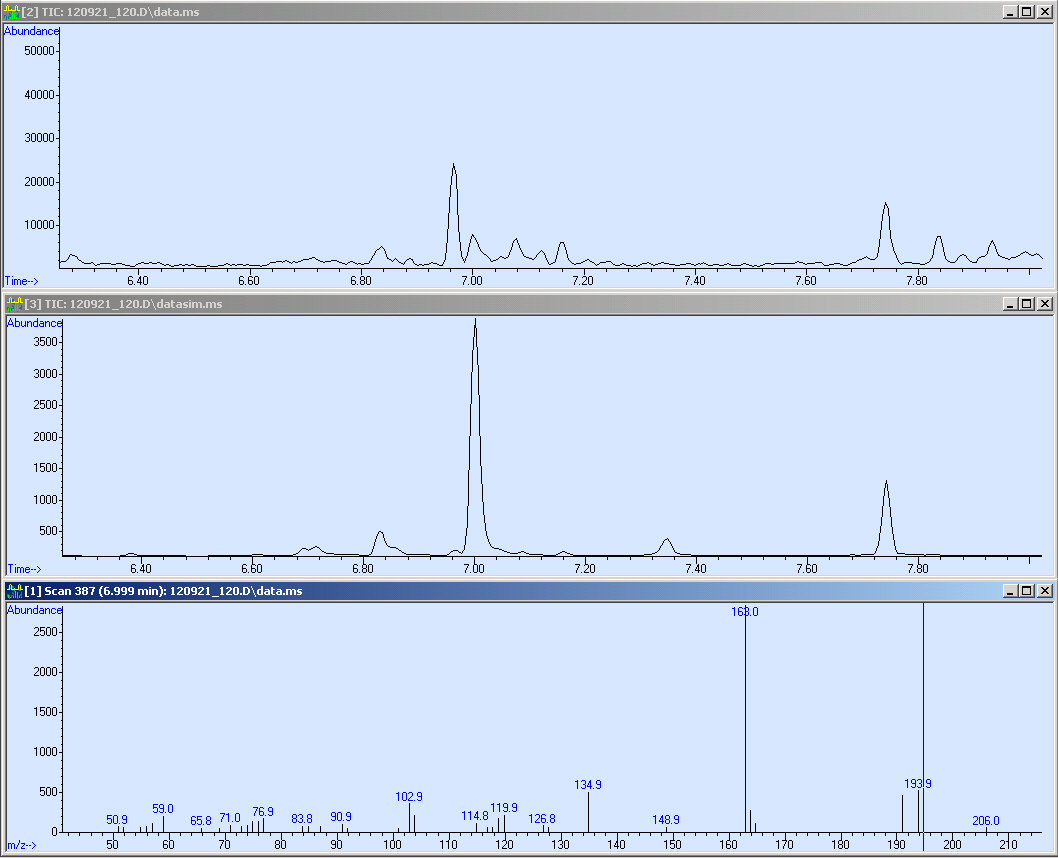
The first sample we measured was OP-2. Simultaneously, it was the proof of principal that our method worked. Here we present the corresponding SCAN, SIM of our sample:
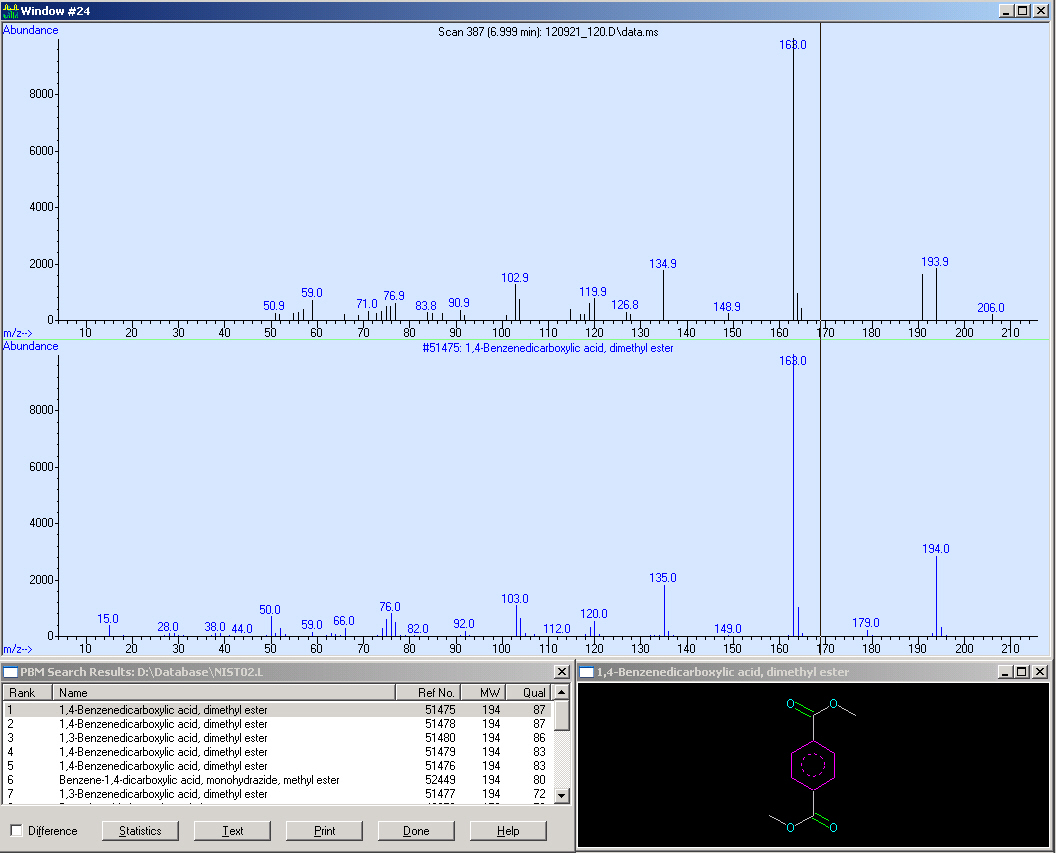
Here we measured a biological sample hence we got a lot of peaks in the SCAN (first window). Surprisingly, in the SIM we got a huge peak at 7 minutes RT. We checked the mass spectrum out of the SCAN (third window) and obtained nearly the same fragmentation pattern we measured in the reference. Due to this, we compared the quality of our spectrum:
Here we can see that our spectrum is nearly identical to the reference spectrum of the TPA-methylester.
This screen-shot shows our SCAN and SIM (zoomed in). Due to the quantification, we need a peak with an sophisticated shape. Fortunately, the peak is perfect for the integration. After the integration we obtained a peak area of 43113.
Notably, we are able to quantify and qualify TPA out of biological sample via GC-MS!
 "
"