Team:TU Darmstadt/Protocols/GC-MS/Results
From 2012.igem.org
Contents |
Sample preparation
We used two methods for our TPA analytics. Therefore we used standard sample preparation protocol:
Purge and Trap
- OP-1 add. 20 µl in 10 ml Water
- OP-2 add. 20 µl in 10 ml Water
- OP-K add. 20 µl in 10 ml Water
TPA short
- OP-1 add. 1ml in vial
- OP-2 add. 1ml in vial
- OP-K add. 1ml in vial
Results
Purge and Trap
Unfortunately, TPA-methyl-ester did not transit into the gas phase. Even though we purged our sample over 10 minutes, TPA-methyl-ester was still part of the solvent.
Here we illustrate the overlay SCAN of OP-1(red), OP-2(green), blank air (black) and the control (blue). Hence we got a lot of peaks, we have to qualify our analyte. Therefore, we extracted an ion-chromatogram with the mass of 163. Due to this we are able to identify our analyte out of the SCAN.
Here we illustrate the ion-chromatogram with the mass of 163. Unfortunately, the peak at 21 RT is column bleeding
Here we illustrate the mass spectrum of the peak at 26 RT. Unfortunately, the peak at 26 RT is column bleeding, too.
TPA Short ATS
Operon 2
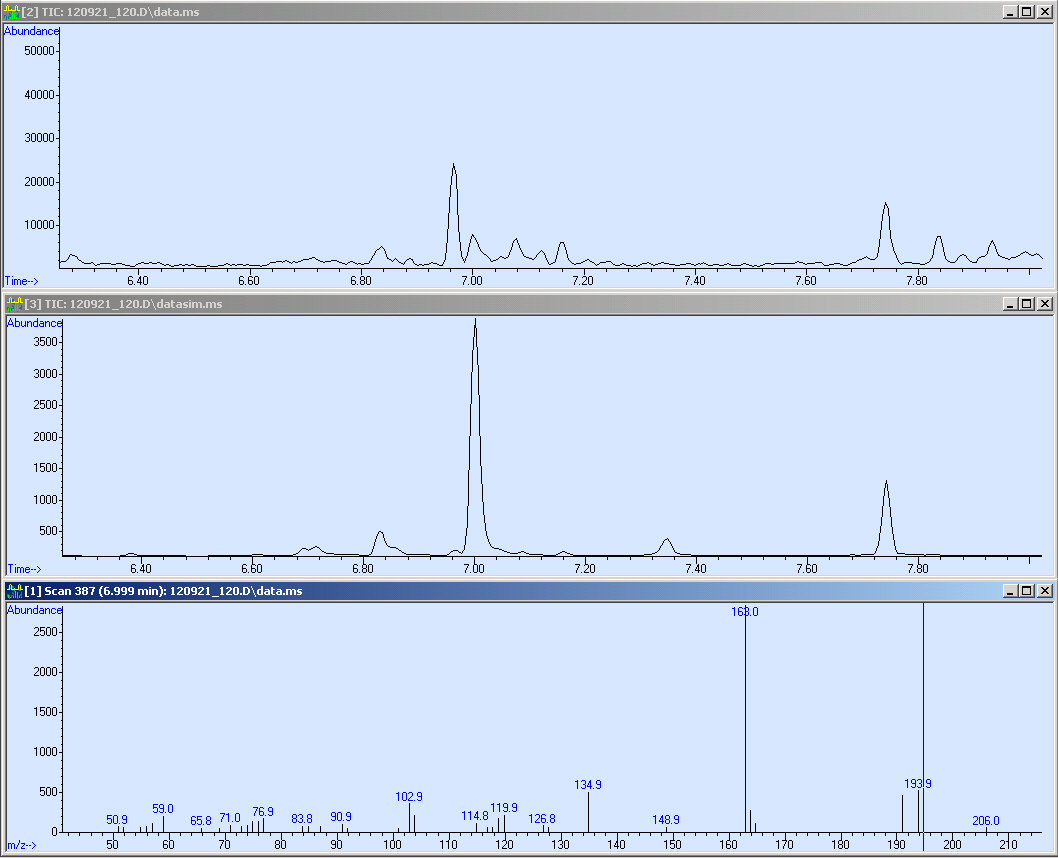
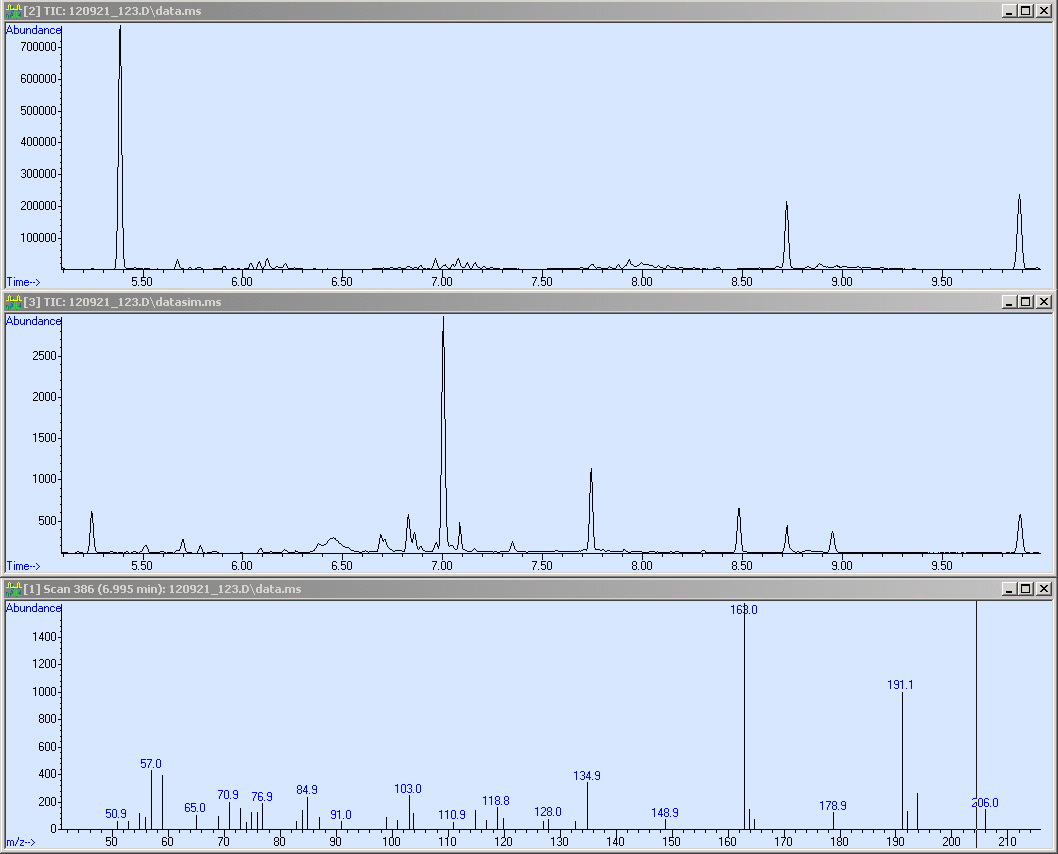
The first sample we measured was Operon 2. Simultaneously, it was the proof of principal that our method worked. Here we present the corresponding SCAN, SIM of our sample:
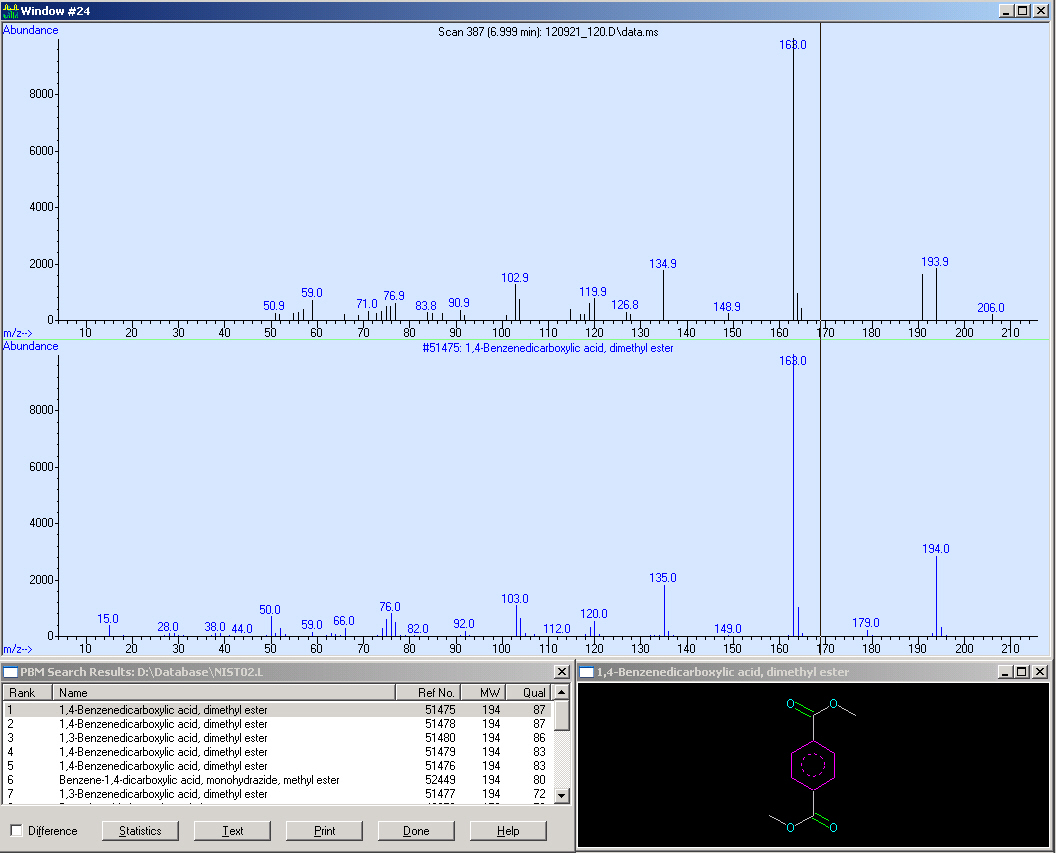
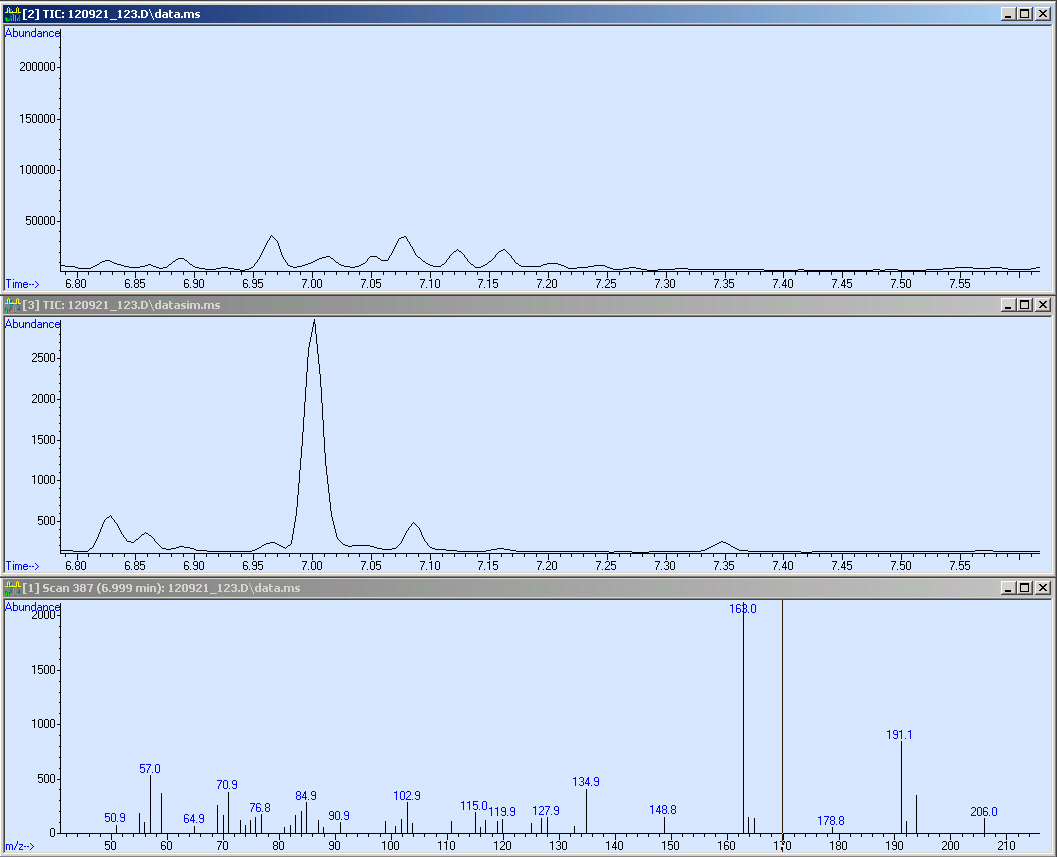
Here we measured a biological sample hence we got a lot of peaks in the SCAN (first window). Surprisingly, in the SIM we got a huge peak at 7 minutes RT. We checked the mass spectrum out of the SCAN (third window) and obtained nearly the same fragmentation pattern as we measured with the reference. Due to this, we compared the quality of our spectrum:
Here we can see that our spectrum is nearly identical to the reference spectrum of the TPA-methylester.
This screen-shot shows our SCAN and SIM (zoomed in). Due to the quantification, we need a peak with an sophisticated shape. Fortunately, the peak is perfect for the integration. After the integration we obtained a peak area of 43113.
Notably, we are now able to quantify and qualify TPA out of biological sample via GC-MS!
Operon 1
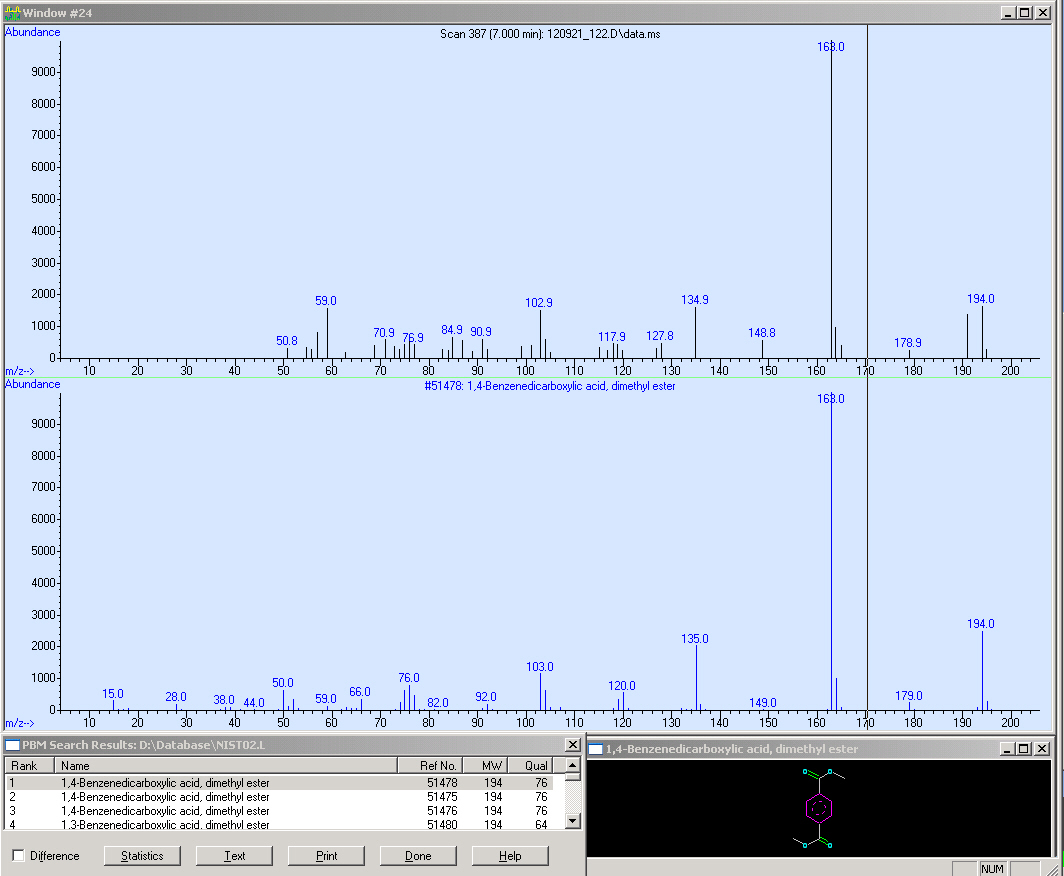
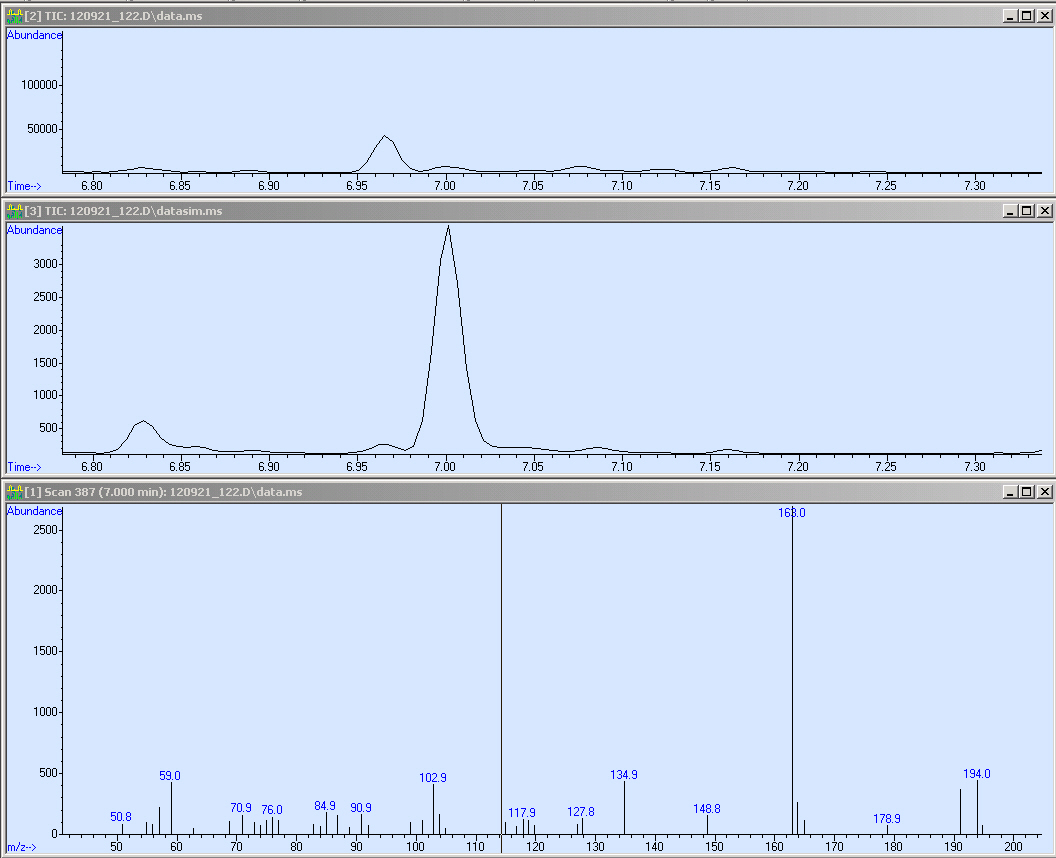
The second sample is Operon 1.
Here you can see the mass spectrum of our molecule as well as the SIM and the SCAN. Hence we are able to reproduce the results from our method (look at the peak at 7 RT)
Here we illustrate the mass spectra of TPA with the corresponding reference spectrum.
After integration of the peak ta 7 RT we obtained an area of 38700.
Control sample
The third sample is our control sample. Notably, the bacteria culture of this sample exposed an 0.85 higher OD than the samples from Operon 1 and 2 thus it will be difficult to compare.
Here we see the SCAN (first window), the SIM (second window) and the third is the mass spectra of the corresponding peak at 7 RT.
After integration of the peak ta 7 RT we obtained an area of 32729
Conclusion
We successfully showed that we are able to qualify and thus quantify TPA-di-methyl-ester via GC-MS. Furthermore, we quantified the operon 1 and operon 2 sample of the metabolism group. Summarising, we obtained the following peak-areas:
- operon-1: 43113
- operon-2: 38700
- control : 32729
Interestingly, we measured a lower concentration of TPA in our control sample. The reason for this might be that the OD of our control sample was higher (0.85) than the OD of our biological samples. Notably, that our untransformed bacteria is able to take TPA up into the periplasma. Nonetheless, due to the the analysis, we measured significant differences in our peak areas. The delta of 4413 digits of Operon-1 to Operon-2 is may be result of a working TPA transport system. For further work we have to measure more samples (biger statistic), measure a control which contains only TPA in the cycl.hexan matrix and using constant ODs by the sample preparations. Moreover, the results have to be compared to HPLC-MS measurements. Unfortunately, the wiki freeze is today, so we are not able to measure again (this time).
 "
"