|
|
| Line 83: |
Line 83: |
| | | | |
| | [[File: UC_Chile-decanalgraph.jpg]] | | [[File: UC_Chile-decanalgraph.jpg]] |
| | + | <html><center><img src="https://static.igem.org/mediawiki/2012/6/6a/Kjasfkjhasdf.jpg" align="right" width="600"></center></html> |
| | | | |
| | While we could measure bioluminescence of the positive LuxBrick E.coli controls, no apparent bioluminescence was seen in our <i>Synechocystis</i> cells. This led us to think that the problem might be the size of the promoter, which if not long enough, would not be able to recruit necessary transcription factors for expression. | | While we could measure bioluminescence of the positive LuxBrick E.coli controls, no apparent bioluminescence was seen in our <i>Synechocystis</i> cells. This led us to think that the problem might be the size of the promoter, which if not long enough, would not be able to recruit necessary transcription factors for expression. |

Construction
Plasmids backbones
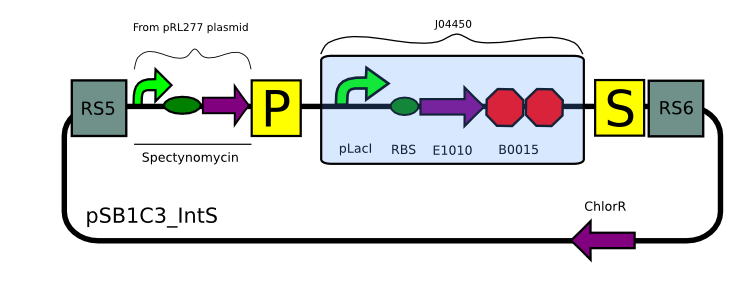
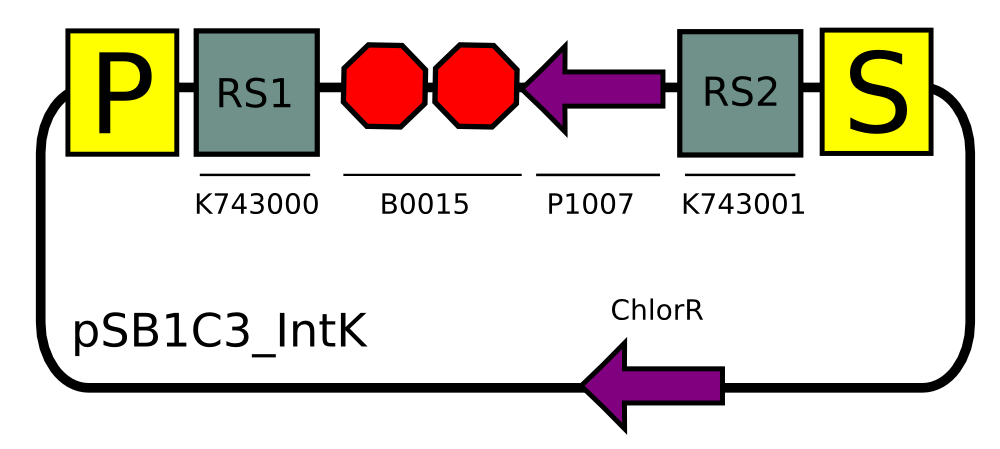
We built two integrative plasmid backbones for Synechocystis PCC. 6803 (see design here)
Plasmids were verified by digestion and then corroborated by sequencing.
Constructs for Synechocystis
Using our plasmid backbones as a starting point, we proceeded to build our constructs for Synechocystis:
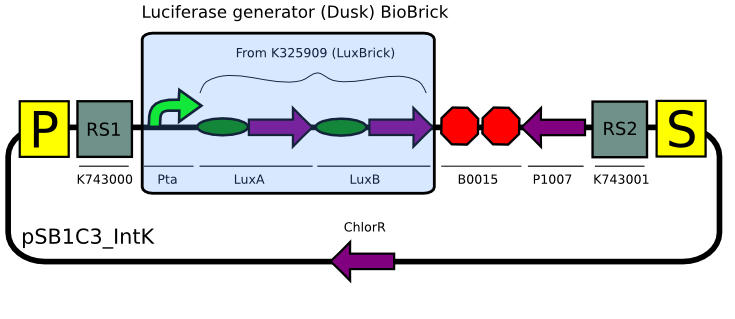
Even though we tried many times (>6) to assemble the substrate production (LuxCDEG) plasmid,we were unable to obtain a correct product. We are currently designing another strategy to build the construct...
Characterization in Synechocystis
Transformation
We used our plasmid backbones to transform Synechocystis PCC. 6803 to verify that our initial design could indeed integrate into Synechocystis's genome.
Transformation with pSB1C3_IntK (BBa_K743015):
Transformation with pSB1C3_IntS (BBa_K743010):
After two weeks, colonies became apparent in the transformation plates for both plasmid backbones while in negative controls (transformations with no DNA) there was complete absence of surviving colonies.
Verification of constructs
We proceeded to verify the integration of the constructs in Synechocystis by doing multiple PCRs to amplify various parts.
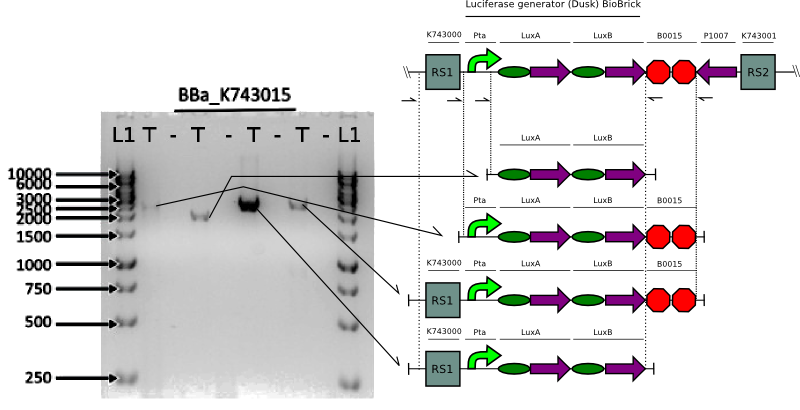
Bioluminescence Assays
We proceeded to check if our Synechocystis strain with the Luciferase genes did indeed produce light in the prescence of exogenous substrate.
Bioluminometer
We measured bioluminescence by adding directly the substrate to the cells and measuring light-output in a Luminometer.
File:UC Chile-decanalgraph.jpg

While we could measure bioluminescence of the positive LuxBrick E.coli controls, no apparent bioluminescence was seen in our Synechocystis cells. This led us to think that the problem might be the size of the promoter, which if not long enough, would not be able to recruit necessary transcription factors for expression.
High-sensitive camera
During the Latin America Jamboree, we had a chat with a couple of judges and a student from Universidad de los Andes, David Olarte, on how to induce Synechocystis with n-decanal.
David sent us some work he had done on luminescence assays in cyanobacteria. Following his methods we finally were able to see light emission, confirming presence of catalytically active luciferase. Thanks David!

Experimental Highlights
- Constructed two functional integration plasmids for Synechocystis PCC. 6803
- Transformed Synechocystis PCC. 6803 with an optimized transformation protocol, available here
- Verified integration of constructs
- Confirmed expression of genes and activity of luciferase through a bioluminescence assay with exogenous substrate.

 "
"