Team:UC Chile/Cyano/Labook/may
From 2012.igem.org
(Difference between revisions)
| Line 47: | Line 47: | ||
<br> | <br> | ||
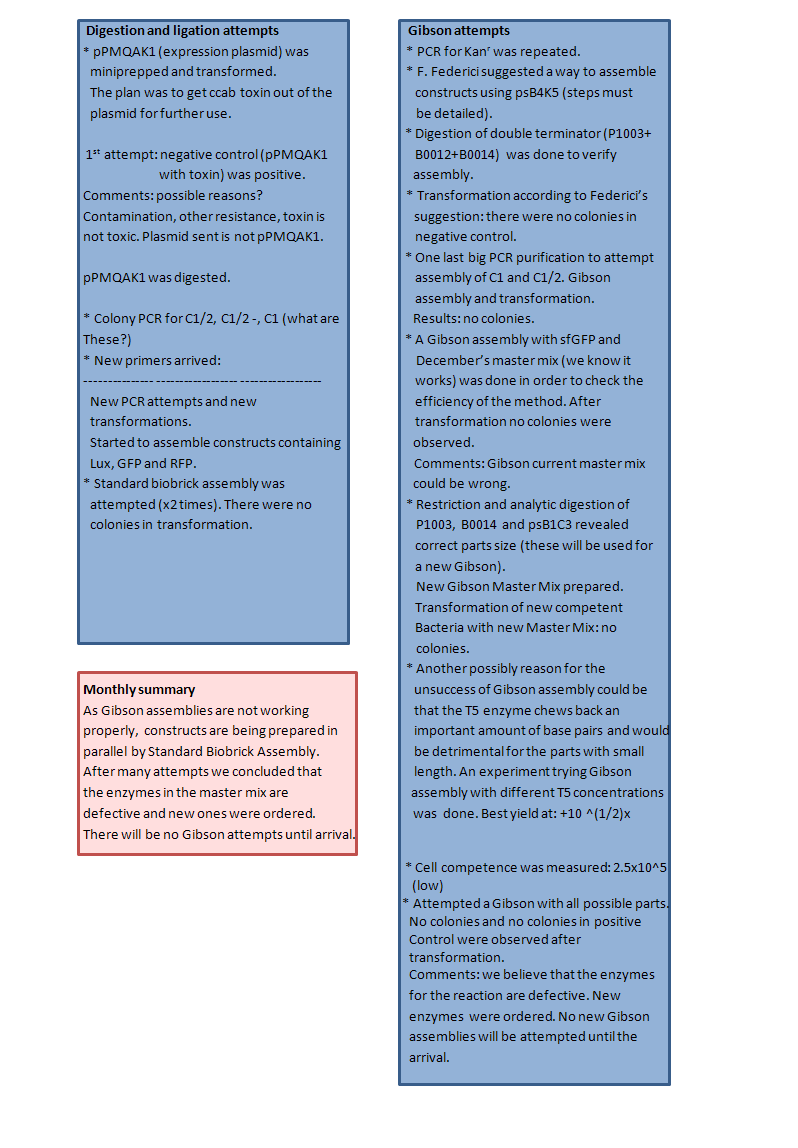
As Gibson assemblies are not working properly, constructs are being prepared in parallel by Standard Biobrick Assembly. After many attempts we concluded that the enzymes in the master mix are defective and new ones were ordered. There will be no Gibson attempts until arrival. | As Gibson assemblies are not working properly, constructs are being prepared in parallel by Standard Biobrick Assembly. After many attempts we concluded that the enzymes in the master mix are defective and new ones were ordered. There will be no Gibson attempts until arrival. | ||
| + | |||
| + | <font size="4">Digestion and Ligation attempts</font> | ||
| + | |||
| + | pPMQAK1 (expression plasmid) wasminiprepped and transformed. | ||
| + | |||
| + | The plan was to get ccab toxin out of the plasmid for further use. | ||
| + | |||
| + | 1st attempt: negative control (pPMQAK1 with toxin) was positive. | ||
| + | |||
| + | Comments: possible reasons? Contamination, other resistance, toxin is not toxic. Plasmid sent is not pPMQAK1. | ||
| + | |||
| + | pPMQAK1 was digested. | ||
| + | |||
| + | Colony PCR for C1/2, C1/2 -, C1 | ||
| + | |||
| + | New primers arrived: | ||
| + | --------------- ------------------ ------------------ | ||
| + | New PCR attempts and new transformations. | ||
| + | |||
| + | Started to assemble constructs containing Lux, GFP and RFP. | ||
| + | |||
| + | Standard biobrick assembly was attempted (x2 times). There were no colonies in transformation. | ||
| + | |||
{{UC_Chilefooter}} | {{UC_Chilefooter}} | ||
Revision as of 21:34, 23 September 2012
 "
"