Team:Arizona State/Notebook
From 2012.igem.org
(Difference between revisions)
Rohit Rajan (Talk | contribs) (→August 15) |
Rohit Rajan (Talk | contribs) |
||
| Line 251: | Line 251: | ||
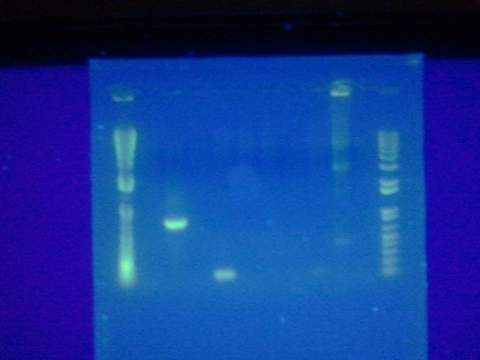
Tried gel for all gel-isolated fragments. Failed. Did not get a band in the 2000 bp region. Only got things below 200 bp.<BR\> | Tried gel for all gel-isolated fragments. Failed. Did not get a band in the 2000 bp region. Only got things below 200 bp.<BR\> | ||
[[Image:ASUiGEM2012_gel081612.jpeg|200px]] | [[Image:ASUiGEM2012_gel081612.jpeg|200px]] | ||
| + | |||
| + | ==August 17== | ||
| + | * GFPT1 sequence confirmed | ||
| + | * Prepared aliquots of GFPT2 minipreps from 8/15/12 for sequencing | ||
| + | * Delivered GFPT2 samples to biodesign for sequencing | ||
| + | |||
| + | ==August 19== | ||
| + | * GFPT2 sequences confirmed | ||
| + | |||
| + | ==August 27== | ||
| + | * Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes | ||
| + | * Made 4mL cultures in LB Amp | ||
| + | |||
| + | ==August 29== | ||
| + | * Discarded GFPT1/2 cultures from 8/27/12 | ||
| + | * Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes | ||
| + | * Made 4mL cultures in LB Amp | ||
| + | * Digested GFPT1/2 with X and S | ||
| + | * Ran a 1% agarose gel with GFPT1/2 digestions | ||
| + | * Cut out inserts and GFPT2 backbone and stored in 4C for gel extraction and tandum repeat assembly experiments | ||
| + | |||
| + | ==August 30== | ||
| + | * Prepared extra glyercol stocks of GFPT1/2 cultures from 8/29/12 | ||
| + | * Miniprepped 3mL of each culture, stored at -20C | ||
| + | |||
| + | ==September 19== | ||
| + | * Set up VF2/VR endpoint PCR for double transform minipreps | ||
| + | ** 1-1, 1-1I, 1-2, 1-2I, 1-3, 1-3I, 2-1, 2-1I, 2-2, 2-2I, 2-3, 2-3I, GFPT1 (positive controls), GFPT2 (positive controls) | ||
| + | * Annealing temp set to 55C for 25 cycles | ||
| + | * Resuspended GFPT1 probe and GFPT2 probe oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C | ||
| + | |||
| + | ==September 25== | ||
| + | * Resuspended pSB1A2 FWD and pSB1A2 REV (amp resistance primers) oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C | ||
| + | * Prepared 1.6uM dilutions (500uL) | ||
| + | * Did endpoint PCR using pSB1A2 primer pair on | ||
| + | ** Topo, Topo IPTG, Topo D168A, Topo D168A IPTG, 1-1, 1-1I, 2-1, 2-1I, GFPT1, GFPT2 | ||
| + | * Did endpoint PCR using VF2/VR primer pair on | ||
| + | ** Topo, Topo IPTG, Topo D168A, Topo D168A IPTG | ||
| + | * Annealing temp 55C for 25 cycles, stored products at -20C | ||
| + | * Made 3mL liquid cultures of the shipping vector (pSB1C3 with RFP insert) in DH5alpha in chloramphenicol resistant LB | ||
| + | * Made 10mL liquid cultures of: | ||
| + | ** Topo in kanamycin | ||
| + | ** topo D168A in kanamycin | ||
| + | ** topo + GFPT1 in kanamycin + ampicillin | ||
| + | ** topo + GFPT2 in kanamycin + ampicillin | ||
| + | * stored @ 37C | ||
==October 1== | ==October 1== | ||
Revision as of 04:31, 3 October 2012
|
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
June 07
- Transformation (LSE)
- Transformed DNA:
- lacZ (well 4:12G, I732019)
- p + lacO (well 1:6G, R0011)
- Cells: neb10beta (donated)
- Protocol from: http://www.neb.com/nebecomm/products/productc3019.asp
- Controls: puc19, no DNA (8 plates)
- Transformed DNA:
June 08
- Transformation results
- puc19: growth
- negative control: no growth
- lacZ, lacO: possible small colonies
- liquid culture in amp media (100 ug / ml):
- no growth of lacZ, lacO
- growth of puc19
June 12
- DH5a Competent Cell Prep
- Streak plated cells on LB no amp plate, let grow overnight
June 13
- Transformation (LSE)
- Transformed DNA:
- lacZ (well 4:12G, I732019)
- p + lacO (well 1:6G, R0011)
- Cells: DH5 alpha (donated)
- Protocol from: http://openwetware.org/wiki/Haynes:Assembly101 (30 minute transformation)
- Controls: puc19, no DNA (8 plates)
- Transformed DNA:
- Transformation (istb4, Abhi)
- Transformed DNA:
- Cells:
- Protocol from:
- Controls:
- DH5a Chemically Competent cell prep
- Grew 2 seed colonies from streak plate in LB no amp
- Grew controls to test for contamination
- Both Seed colonies grew, no contamination present
June 14
- Competent cell prep
- Prepared CaCl2 buffer solution and CaCl2 glycerol buffer solution
- Grew seed colony in 400mL LB no amp
June 15
- Competent cell prep
- Centrifuged falcon test tubes containing liquid colonies
- Resuspended in CaCl2 buffer solution and incubated for 15 mins
- Centrifuged and resuspended in CaCl2 glycerol buffer solution
- Chilled overnight
June 16
- Competent cell prep
- Aliquotted 200uL into test tubes
- Stored in -80C
June 17
- Streak plated prepared competent cells on LB no amp plate
- Colonies observed
June 19
- Transformation (LSE)
- Transformed DNA:
- T7 promoter BBa_I712074
- Constitutive promoter BBa_J23102
- Cells: DH5 alpha (donated)
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Controls: puc19, no DNA
- Plated 2 copies of each (100 ul, 250 ul) on LB amp plates.
- Transformed DNA:
- Made 50 LB Amp plates.
June 20
- Plated negative control on LB Amp plate
- Liquid cultures of T7 promoter and constitutive promoter
- Transformation (LSE)
- Transformed DNA:
- RBS (well 1:1H BBa_B0030)
- TetR GFP (well 2:8A Part:BBa_I13522)
- Cells: DH5 alpha (donated)
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Controls: puc19, no DNA
- Transformed DNA:
June 21
- Made Liquid Cultures of E.coli transformed with RBS B0030
- Made Liquid Cultures of E.coli transformed with TetR GFP
- miniprepped and nanodropped T7 promoter BBa_I712074 and Constitutive promoter BBa_J23102 liquid cultures
- liquid cultures:
- RBS1
- RBS2 (duplicate_
- GFP1
- puc19
- negative controls
- 5 ml LB amp
- overnight cultures
- replated GFP1 & 2 (duplicates)
- Nanodropped plasmid DNA samples
- Constitutive promoter 1: __ng/uL
- Constitutive promoter 2: __ng/uL
- T7 promoter 1: __ng/uL
- T7 promoter 2: __ng/uL
June 22
- Miniprepped liquid cultures: RBS (well 1:1H BBa_B0030) and TetR GFP (well 2:8A Part:BBa_I13522)
- Picked colonies:
- 1 colony from double terminator (dt1) plate
- 1 colony from t7 polymerase (pol1) plate
- 1 colony from puc19 plate (positive control)
- 1 colony from dh5a plate (negative control)
- started liquid cultures of each colony (5 mL LB amp each)
June 26
- Transformation:
- Transformed DNA:
- double terminator (B0017, 2:6K)
- T7 RNA polymerase (I715038, 2:15C)
- puc19, negative control
- Protocol from: http://tools.invitrogen.com/content/sfs/manuals/subcloningefficiencydh5alpha_man.pdf (30 minute transformation)
- Cells: dh5a
- Transformed DNA:
June 27
- 6-26 transformation results:
- Controls correct
- 2x terminator: ~19 colonies
- RNA pol: 1 colony
- Liquid cultures including controls
June 28
- Miniprepped double terminator (B0017, 2:6K) and T7 RNA polymerase (I715038, 2:15C) liquid cultures
July 2
- Cleaned up liquid waste
- Made SOB media
- Finalized oligos for magainin construct
July 3
- Autoclaved SOB media
- Added glucose to make SOC media
- Nanodropped double terminator (B0017, 2:6K) [DT1: 24.5, DT2: 29.6] and T7 RNA polymerase (I715038, 2:15C) [P1: 64.6, P2: 55.3] liquid cultures
July 24
- Plasmids arrived courtesy of University of Pennsylvania School of Medicine
- pET29a vectors containing coding sequence for Topoisomerase mutants CSCS and CSCS D168A described here
July 25
- Tranformed CSCS topo 0 plasmid and CSCS D168A topo into DH5a Turbo cells (with neg control and Puc19 neg control)
- Plated on Kanamycin plates
July 26
- Picked colonies colonies from Topo 0 and Topo D168A and grew liquid cultures in Kan media
July 27
- Miniprepped liquid colonies and nanodropped.
- Plasmid concentrations
- Topo O:
- Topo D168A1:
- Topo D168A2:
July 30
- Prepared Kan Media and Kan Plates
July 31
- PCR amplified polylinker sequence of Topo plasmid with Promega GoTaq protocol
- Used pET29a upstream forward primer and T7 terminator reverse primer
August 3
- Submitted pET29a Topoisomerase plasmid to Biodesign for sequencing
- Resuspended GFPT1 and GFPT2 oligos with molecular grade (nuclease-free) H2O.
- Final Concentration 100uM
- (gfpt1 top1, gfpt2 top1, gfpt1 top2, gftp2 top2, gfpt1 bot1, gfpt2 bot1, gfpt1 bot2, gfpt2 bot2)
- (3uL of each oligo + 2uL 10x annealing buffer, 6uL molecular grade H2O. 20uL Reactions)
- Final Concentration 100uM
- Heated for 5 minutes at 100C. Let cool to room temperature on the heating block, stored at -20C.
- Digested BBa_I13522 with XbaI and PstI.
- Attempted ligating annealed oligos into a digested plasmid from Ryan (realized it was cut with E and P).
August 6
- Annealed oligos for GFPT1 and GFPT2 (target probes)
- Ligated oligos with digested GFP plasmid (BBa_I13522)
- Transformed into competent DH5alpha
- Added SOC and incubated at 37C for 15 minutes.
- Plated on amp treated plates.
August 7
- Only one colony on each plate (both were white)
- Picked colonies and started 5mL LB amp cultures of each, stored at 37C
- Stored plates in 37C
August 8
- Picked the colonies again and started new liquid cultures (5mL LB amp).
- Discarded cultures for 8/7/12
August 9
- Miniprepped 3mL of each 8/8/12 culture and nanodropped:
- gfpt1 - 155 ng/uL
- gfpt2 - 114 ng/uL
- Digested gfpt1 and gfpt2 with X and P
- Ran on a 1% agarose gel with the digested GFP plasmid
- Made glyercol stocks with aliquot of the remaining liquid cultures
August 10
- PCR of Alpha-4, 1-omega, omega, and alpha fragments using corrected primers
August 13
- Ran gel of split beta gal fragments. Confirmed 3 out of the 4 fragments except for the alpha-4 fragment.
- Prepared sequencing samples
- Sample w/ Primer:
- GFPT1 w/ VF2 GFPT1 w/ VR GFPT2 w/ VF2 GFPT2 w/ VR
- 200ng of DNA + 16 pmol of primer
- Annealed oligos again GFPT1/2
- Repeated ligation of oligos with digested GFP plasmid (BBa_I13522)
- Followed Haynes assembly protocol instead of standard DH5alpha protocol. (http://openwetware.org/wiki/Haynes:Assembly101)
- Transformed ligations into competent DH5alpha
- Plated on amp treated plates
August 14
- Took pictures of plates
- Green-white screened plates
- Picked 4 white colonies from each of gfpt1/2 plates
- Made 5mL LB amp cultures of each colony
- Delivered GFPT1/2 dna samples to biodesign for sequencing (samples from 8/13/12)
- Assembled magainin insert via Overlapping oligo assembly
- Digested pUC 19 plasmid with EcoRI and PstI
- Transformed Magainin insert into digested pUC 19 plasmid. Failed. Probably too much X-gal on plate.
Ran gel for the beta gal alpha-4 fragment. Failed. Fragment not in the correct size-band.
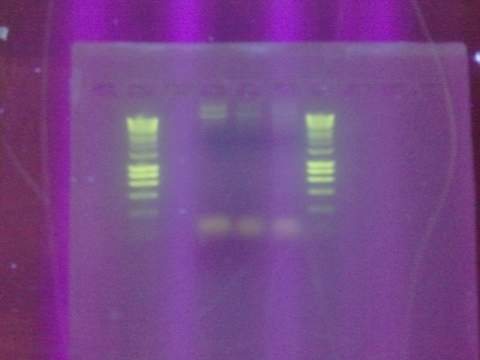
August 15
- [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/topo0_T7Term.seq Topo 0], [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo1_T7Term.seq D168A Topo1], and [http://129.219.2.10/home2/dnalims/fragment/32/21/pfasta/D168Atopo2_T7Term.seq D168A Topo2] sequence results
- Miniprepped 3mL of each liquid culture of GFPT1/2
- Prepared glycerol stocks using 100uL of each liquid culture
- Nanodropped samples:
- GFPT1-1 - 172.6 ng/uL
- GFPT1-2 - 203.7 ng/uL
- GFPT1-3 - 197.4 ng/uL
- GFPT1-4 - 178.9 ng/uL
- GFPT2-1 - 107.3 ng/uL
- GFPT2-2 - 131.2 ng/uL
- GFPT2-3 - 145.5 ng/uL
- GFPT2-4 - 172.0 ng/uL
August 16
- Replated magainin insert + plasmid into the grid. Failed. All blue colonies meaning that no insert.
Tried gel for all gel-isolated fragments. Failed. Did not get a band in the 2000 bp region. Only got things below 200 bp.
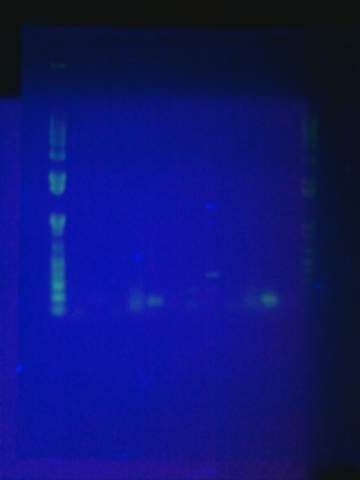
August 17
- GFPT1 sequence confirmed
- Prepared aliquots of GFPT2 minipreps from 8/15/12 for sequencing
- Delivered GFPT2 samples to biodesign for sequencing
August 19
- GFPT2 sequences confirmed
August 27
- Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes
- Made 4mL cultures in LB Amp
August 29
- Discarded GFPT1/2 cultures from 8/27/12
- Revived GFPT1 (from 8/9/12) and GFPT2 (2-2 from 8/15/12) cultures from glycerol scrapes
- Made 4mL cultures in LB Amp
- Digested GFPT1/2 with X and S
- Ran a 1% agarose gel with GFPT1/2 digestions
- Cut out inserts and GFPT2 backbone and stored in 4C for gel extraction and tandum repeat assembly experiments
August 30
- Prepared extra glyercol stocks of GFPT1/2 cultures from 8/29/12
- Miniprepped 3mL of each culture, stored at -20C
September 19
- Set up VF2/VR endpoint PCR for double transform minipreps
- 1-1, 1-1I, 1-2, 1-2I, 1-3, 1-3I, 2-1, 2-1I, 2-2, 2-2I, 2-3, 2-3I, GFPT1 (positive controls), GFPT2 (positive controls)
- Annealing temp set to 55C for 25 cycles
- Resuspended GFPT1 probe and GFPT2 probe oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C
September 25
- Resuspended pSB1A2 FWD and pSB1A2 REV (amp resistance primers) oligos in molecular grade H2O (Final concentration: 100uM), stored at -20C
- Prepared 1.6uM dilutions (500uL)
- Did endpoint PCR using pSB1A2 primer pair on
- Topo, Topo IPTG, Topo D168A, Topo D168A IPTG, 1-1, 1-1I, 2-1, 2-1I, GFPT1, GFPT2
- Did endpoint PCR using VF2/VR primer pair on
- Topo, Topo IPTG, Topo D168A, Topo D168A IPTG
- Annealing temp 55C for 25 cycles, stored products at -20C
- Made 3mL liquid cultures of the shipping vector (pSB1C3 with RFP insert) in DH5alpha in chloramphenicol resistant LB
- Made 10mL liquid cultures of:
- Topo in kanamycin
- topo D168A in kanamycin
- topo + GFPT1 in kanamycin + ampicillin
- topo + GFPT2 in kanamycin + ampicillin
- stored @ 37C
October 1
- Minipreps of:
- J61011 + S + L + ALPHA 2B
- J61100 + S + L + ALPHA 2B
- J61100 + S + L + ALPHA 2C
- PLUX2 + RBS + ALPHA 1A
- PLUX2 + RBS + ALPHA 1A (2)
- PLUX + S + L + ALPHA 2C
- J61101 + S + L + ALPHA 1A
- J61101 + S + L + ALPHA 1A (2)
- PLUX2 + RBS + S + L + ALPHA 2B
- PLUX2 + RBS + S + L + ALPHA 2B (2)
- J61101 + S + L + ALPHA 2C
- PLUX + S + L + ALPHA 2B
- Ran Twice when finding concentrations:
- J61101 + S + L + ALPHA 1A
- J61101 + S + L + ALPHA 2B
- Restricted all the above and:
- Strep + Linker + OMEGA
- Strep + Linker-4 + OMEGA
- with:
- X+P
- Ligated into pSB1C3 shipping vector and transformed into BL21(DE3) cells.
- Prepared all 14 above samples for sequencing.
 "
"