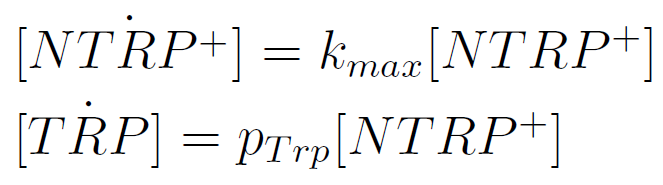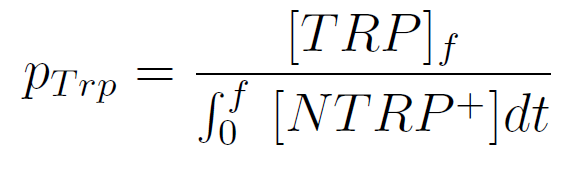Team:Buenos Aires/Results/BBsTesting
From 2012.igem.org
(→Experimental determination of strains death rate) |
m (→Results) |
||
| (83 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
= After the Jamboree! = | = After the Jamboree! = | ||
| - | When we returned from the Latin America Jamboree we focused all our efforts on completing the neccesary transformations to test our devices and our | + | |
| + | DEVICE TESTING | ||
| + | |||
| + | https://2012.igem.org/Team:Buenos_Aires/Results/DevicesTesting | ||
| + | |||
| + | |||
| + | SYNECO TESTING | ||
| + | |||
| + | https://2012.igem.org/Team:Buenos_Aires/Results/SynEcoTesting | ||
| + | |||
| + | |||
| + | |||
| + | When we returned from the Latin America Jamboree we focused all our efforts on completing the neccesary transformations to test our devices and our project as a whole. | ||
We devided this task in three sections | We devided this task in three sections | ||
| + | |||
== Week 1&2 : Yeast expression vectors & Transformations == | == Week 1&2 : Yeast expression vectors & Transformations == | ||
| Line 17: | Line 30: | ||
|[[File:BsAs2012-plasmid-BPCM185.gif|340px]] | |[[File:BsAs2012-plasmid-BPCM185.gif|340px]] | ||
|} | |} | ||
| - | |||
| - | |||
The cloning we did was: | The cloning we did was: | ||
| Line 56: | Line 67: | ||
# Purification of digested vectors (pCM185; pCM182; pEG202) | # Purification of digested vectors (pCM185; pCM182; pEG202) | ||
# Ligation of vectors and devices according the anterior table (T4 ligase protocol, overnight) | # Ligation of vectors and devices according the anterior table (T4 ligase protocol, overnight) | ||
| - | # Transformation of E. Coli DH5a with the ligation products. Bacterias were plated on LB-agar with | + | # Transformation of E. Coli DH5a with the ligation products. Bacterias were plated on LB-agar with Ampicillin, and incubated over night at 37 °C. |
| - | # Colonies were used for liquid cultures (LB + | + | # Colonies were used for liquid cultures (LB + Ampicillin) and minipreps were made. |
# Constructions (vector + insert) were checked by digestion with restriction enzymes, and 1%-agarose gel (1kb and 100bp as markers). | # Constructions (vector + insert) were checked by digestion with restriction enzymes, and 1%-agarose gel (1kb and 100bp as markers). | ||
| Line 65: | Line 76: | ||
# TCY3128: W303, bar1-, leu2:: Pprm1-CFP 405 | # TCY3128: W303, bar1-, leu2:: Pprm1-CFP 405 | ||
| - | |||
| - | {| | + | |
| - | | Name | + | We got the following '''transformed strains''': |
| - | | | + | |
| - | | Plasmid | + | {| |
| + | | | ||
| + | {| style="width:50%" | ||
| + | | rowspan="3" | [[File:BsAs2012-icono-Cepa1.jpg | 200px]] | ||
| + | ! scope="row" style="background: #7ac5e8"| Name | ||
| + | | '''YFP_TRPa_TRPZipper2''' | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3081 (YFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pCM182 (TRPa) + <partinfo>BBa_K792010</partinfo> (TRPZipper2) | ||
| + | |} | ||
| + | | | ||
| + | {| style="width:50%" | ||
| + | | rowspan="3" | [[File:BsAs2012-icono-Cepa2.jpg | 200px]] | ||
| + | ! scope="row" style="background: #7ac5e8"| Name | ||
| + | | '''YFP_TRPa_PoliWb''' | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3081 (YFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pCM182 (TRPa) + <partinfo>BBa_K792012</partinfo> (PoliWb) | ||
| + | |} | ||
|} | |} | ||
| - | + | {| | |
| - | + | | | |
| - | + | {| style="width:50%" | |
| - | pCM185 (TRPb) + BBa_K792010 (TRPZipper2) | + | | rowspan="3" | [[File:BsAs2012-icono-Cepa3.jpg | 200px]] |
| - | pCM185 (TRPb) + BBa_K792012 (PoliWb) | + | ! scope="row" style="background: #7ac5e8"| Name |
| - | pEG202 (HIS) + BBa_K792009 (PoliHa) | + | | '''YFP_TRPb_TRPZipper2''' |
| - | pEG202 (HIS) + BBa_K792011 (PoliHb) | + | |- |
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3081 (YFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pCM185 (TRPb) + <partinfo>BBa_K792010</partinfo> (TRPZipper2) | ||
| + | |} | ||
| + | | | ||
| + | {| style="width:50%" | ||
| + | | rowspan="3" | [[File:BsAs2012-icono-Cepa4.jpg | 200px]] | ||
| + | ! scope="row" style="background: #7ac5e8"| Name | ||
| + | | '''YFP_TRPb_PoliWb''' | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3081 (YFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pCM185 (TRPb) + <partinfo>BBa_K792012</partinfo> (PoliWb) | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | {| | ||
| + | | | ||
| + | {| style="width:50%" | ||
| + | | rowspan="3" | [[File:BsAs2012-icono-Cepa6.jpg | 200px]] | ||
| + | ! scope="row" style="background: #7ac5e8"| Name | ||
| + | | '''CFP_HIS_PoliHa''' | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3128 (CFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pEG202 (HIS) + <partinfo>BBa_K792009</partinfo> (PoliHa) | ||
| + | |} | ||
| + | | | ||
| + | {| style="width:50%" | ||
| + | | rowspan="3" | [[File:BsAs2012-icono-Cepa5.jpg | 200px]] | ||
| + | ! scope="row" style="background: #7ac5e8"| Name | ||
| + | | '''CFP_HIS_PoliHb''' | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Strain | ||
| + | |TCY3128 (CFP) | ||
| + | |- | ||
| + | ! scope="row" style="background: #CEE3F6"|Plasmid | ||
| + | |pEG202 (HIS) + <partinfo>BBa_K792011</partinfo> (PoliHb) | ||
| + | |} | ||
| + | |} | ||
== Week 3: Synthetic Ecology Characterization == | == Week 3: Synthetic Ecology Characterization == | ||
| Line 91: | Line 171: | ||
In order to characterize the devices that we used to transform our strains we came up with the series of assays that we describe below. | In order to characterize the devices that we used to transform our strains we came up with the series of assays that we describe below. | ||
| + | |||
| + | |||
=== Devices characterization === | === Devices characterization === | ||
| Line 96: | Line 178: | ||
==== Secretion Rate of Trp as a function of culture growth ==== | ==== Secretion Rate of Trp as a function of culture growth ==== | ||
| - | The first step was to actually check if the construct works: do the transformed yeast strains - with the | + | The first step was to actually check if the construct works: do the transformed yeast strains - with the tryptophan devices- actually secrete tryptophan into the medium? |
To test this we used the following strains: | To test this we used the following strains: | ||
| - | + | {| | |
| - | + | | | |
| - | + | {| | |
| - | + | |- | |
| - | + | |[[File:BsAs2012-icono-Cepa3.jpg|200px]] | |
| - | + | |[[File:BsAs2012-icono-Cepa4.jpg|200px]] | |
| - | + | |[[File:BsAs2012-icono-YFP-185.jpg|200px]] | |
| + | |- align="center" | ||
| + | |YFP_TRPb_TRPZipper2 | ||
| + | |YFP_TRPb_PolyWb | ||
| + | |YFP_TRPb | ||
| + | |} | ||
| + | | rowspan="2" | | ||
| + | {| | ||
| + | | [[File:BsAs2012-icono-YFP.jpg | 200px]] | ||
| + | |- align="center" | ||
| + | |YFP | ||
| + | |} | ||
| + | |||
| + | |- | ||
| + | | | ||
| + | {| | ||
| + | |- | ||
| + | |[[File:BsAs2012-icono-Cepa1.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-Cepa2.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-YFP-182.jpg | 200px]] | ||
| + | |- align="center" | ||
| + | |YFP_TRPa_TRPZipper2 | ||
| + | |YFP_TRPa_PolyWb | ||
| + | |YFP_TRPa | ||
| + | |} | ||
| + | |||
| + | |} | ||
| + | |||
| - | + | ===== Protocol ===== | |
*We started 5ml cultures with 3 replica until they reached exponential phase, overnight, using a -T medium. | *We started 5ml cultures with 3 replica until they reached exponential phase, overnight, using a -T medium. | ||
| Line 115: | Line 224: | ||
*We measured the Trp signal for each culture medium using the spectrofluorometer. | *We measured the Trp signal for each culture medium using the spectrofluorometer. | ||
| + | NOTE: The fluorescence meausurements taken for the Tryptophan in the medium, take into account both the aminoacid secreted by the device and the one diffused. | ||
| - | We used a simple model to measure the | + | Therefore the secretion rates calculated will be higher than the actual ones. We used an empty plasmid as control to study tryptophan diffusion. |
| + | |||
| + | We used a simple model to measure the secretion rate for all the strains. Since the cultures are in exponential phase, we take | ||
[[File:BsAs2012-eqTrp1.jpg | 225px]] | [[File:BsAs2012-eqTrp1.jpg | 225px]] | ||
| Line 124: | Line 236: | ||
[[File:BsAs2012-eqTrp2.jpg | 225px]] | [[File:BsAs2012-eqTrp2.jpg | 225px]] | ||
| - | + | ===== Results ===== | |
| - | + | Next we show the average OD for each strains used, needed to calculate the secretation rates of Trytophan. | |
| - | + | {| | |
| + | [[File:BsAs2012Odvstimea4.jpg | 350px]] | ||
| + | | | ||
| + | [[File:BsAs2012Odvstimeab.jpg | 350px]] | ||
| + | |} | ||
| + | Figure X. check | ||
| - | [[File: | + | [[File:BsAs2012rate2.jpg| 350px]] |
| + | |||
| + | Figure X. check | ||
==== Tryptophan secretion at increasing histidine concentrations ==== | ==== Tryptophan secretion at increasing histidine concentrations ==== | ||
| - | We asked ourselves which was the dependance of tryptophan secretion on the amount of histidine in medium. We carried on this test in order to determine | + | We asked ourselves which was the dependance of tryptophan secretion on the amount of histidine in medium. We carried on this test in order to determine whether the secretion of tryptophan depends on the concentration of another aminoacid in the media, such as histidine and what would be the necessary amount of histidine in medium for the start of our system. |
In this experiment we used strains: | In this experiment we used strains: | ||
| - | |||
| - | - YFP_TRPb_PolyWb | + | {| |
| + | |- | ||
| + | |[[File:BsAs2012-icono-Cepa3.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-Cepa4.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-YFP-185.jpg|200px]] | ||
| + | |- align="center" | ||
| + | |YFP_TRPb_TRPZipper2 | ||
| + | |YFP_TRPb_PolyWb | ||
| + | |YFP_TRPb (control) | ||
| + | |} | ||
| - | |||
| - | + | ||
| + | |||
| + | |||
| + | |||
| + | ===== Protocol ===== | ||
# Starters of each strain used were grown over night in -T media, at 30 °C in shaker. | # Starters of each strain used were grown over night in -T media, at 30 °C in shaker. | ||
| Line 152: | Line 282: | ||
# We left the cultures in shaker at 30ºC for 5 hours. After that time, we measured the final OD reached by cultures with the use of a spectrophotometer and the amount of tryptophan present in medium with a spectrofluorometer. | # We left the cultures in shaker at 30ºC for 5 hours. After that time, we measured the final OD reached by cultures with the use of a spectrophotometer and the amount of tryptophan present in medium with a spectrofluorometer. | ||
| - | + | ===== Results ===== | |
| - | [[File: | + | [[File: BsAs2012TrpvHisvv.jpg|350px]] |
| - | + | ||
| - | + | ||
=== Strain characterization === | === Strain characterization === | ||
==== Experimental determination of strains death rate==== | ==== Experimental determination of strains death rate==== | ||
| - | We set out to determine how long can auxotroph cells[link] survive in media that lacks both Trytophan and Histidine. These values are the '''death''' parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis | + | We set out to determine how long can auxotroph cells[link] survive in media that lacks both Trytophan and Histidine. These values are the '''death''' parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis for simplicity but now we would like to test whether this approximation is accurate. |
Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our '' system's feasability''. | Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our '' system's feasability''. | ||
| - | + | ===== Protocol ===== | |
| - | *We set cultures of the two | + | For this experiment we used |
| + | {| | ||
| + | |- | ||
| + | |[[File:BsAs2012-icono-YFP.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-CFP.jpg|200px]] | ||
| + | |- align="center" | ||
| + | |YFP Strain | ||
| + | |CFP Strain | ||
| + | |} | ||
| + | |||
| + | *We set cultures of the two auxotroph strains without being transformed (YFP and CFP) in medium –HT at an initial OD of 0.01. | ||
*Each day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain. | *Each day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain. | ||
| - | + | ===== Result ===== | |
| - | + | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
! scope="row" style="background: #7ac5e8" |Strain | ! scope="row" style="background: #7ac5e8" |Strain | ||
| Line 216: | Line 355: | ||
|} | |} | ||
| - | Table: Number of colonies counted per plate. | + | '''Table:''' Number of colonies counted per plate. |
| + | |||
| + | We expect to see a decrease in the number of colonies - because of cell death. We found that this was not the case, in the experiment's time lapse. However we observed that the size of the colonies was smaller everyday as can be seen in the following pictures. | ||
| + | |||
| + | [[File:Bsas2012kdeathcells.png| 500px]] | ||
| + | |||
| + | |||
| + | We can infer from this data that though they have not died, they may have enter into a '''...Alan state'''. In this way cells can survive for a period of time in media defficient in amino acid (at least, during the time course of our experiment), but grow slower. Probably this would require more time than 3 days to observe significative cell dying. | ||
| - | |||
{| | {| | ||
| Line 229: | Line 374: | ||
==== Growth dependence on the Trp and His concentration==== | ==== Growth dependence on the Trp and His concentration==== | ||
| - | An important thing to characterize | + | An important thing in order to characterize the system is the dependence of the growth rate on the culture with the concentration of the crossfeeding aminoacids, tryptophane (Trp) and histidine (His). |
| - | This would allow us to estimate one of the parameters used in our model (EC50, the amount of aminoacid at which the culture reaches half of the maximum growth). | + | This would allow us to estimate one of the parameters used in our model (the EC50, which is the amount of aminoacid at which the culture reaches half of the maximum growth). |
===== Protocol===== | ===== Protocol===== | ||
| - | 1. | + | 1. For this experiment we would use strains YFP and CFP (non-transformed, without devices). |
We started cultures of 5 ml of initial OD: 0.025 for: | We started cultures of 5 ml of initial OD: 0.025 for: | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | {| class="wikitable" | ||
| + | | Strain YFP at the following media [Trp] = 1x; 0.5x; 0.25x; 0.125x; 0.0625x, 0.03125x | ||
| + | |- | ||
| + | | Strain CFP at the following media [His] = 1x; 0.5x; 0.25x; 0.125x; 0.0625x, 0.03125x | ||
| + | |- | ||
| + | | Strain YFP at Synthetic Complete media | ||
| + | |- | ||
| + | | Strain CFP at Synthetic Complete media | ||
| + | |} | ||
| + | |||
2. Cultures would be left overnight (12 hs) at 30°C with agitation and we measured OD reached with the use of spectrophotometer. | 2. Cultures would be left overnight (12 hs) at 30°C with agitation and we measured OD reached with the use of spectrophotometer. | ||
===== Results ===== | ===== Results ===== | ||
| - | To be done | + | To be done with our strains soon. |
=== Coculture of transformed strains === | === Coculture of transformed strains === | ||
| Line 257: | Line 404: | ||
We designed an assay to do so; following the growth of these strains: | We designed an assay to do so; following the growth of these strains: | ||
| + | |||
| + | {| | ||
| + | |- | ||
| + | |[[File:BsAs2012-icono-CFP-202.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-YFP-185.jpg|200px]] | ||
| + | |- align="center" | ||
| + | |CFP_His | ||
| + | |YFP_TRPb | ||
| + | |} | ||
| + | |||
| + | |||
{| | {| | ||
| Line 334: | Line 492: | ||
| 384 wells plate to be used for epifluorescence microscope. | | 384 wells plate to be used for epifluorescence microscope. | ||
|} | |} | ||
| - | |||
==== Results ==== | ==== Results ==== | ||
| + | |||
| + | [[File:BsAs2012coculture1.jpg | 600px]] | ||
| + | |||
| + | [[File:BsAs2012coculture2.jpg| 600px]] | ||
Latest revision as of 03:55, 27 October 2012

Contents |
After the Jamboree!
DEVICE TESTING
https://2012.igem.org/Team:Buenos_Aires/Results/DevicesTesting
SYNECO TESTING
https://2012.igem.org/Team:Buenos_Aires/Results/SynEcoTesting
When we returned from the Latin America Jamboree we focused all our efforts on completing the neccesary transformations to test our devices and our project as a whole.
We devided this task in three sections
Week 1&2 : Yeast expression vectors & Transformations
In order to construct the yeast expression plasmids we choosed 3 vectors, 2 with a tryptophan marker and 1 with an histidine marker:
- pCM182/5, which are centromeric plasmids with TRP1 marker, and with a doxycycline repressible promoter [Gari et al 1996].
- pEG202, with a 2 ori, HIS3 marker and a constitutive promoter (PADH1).
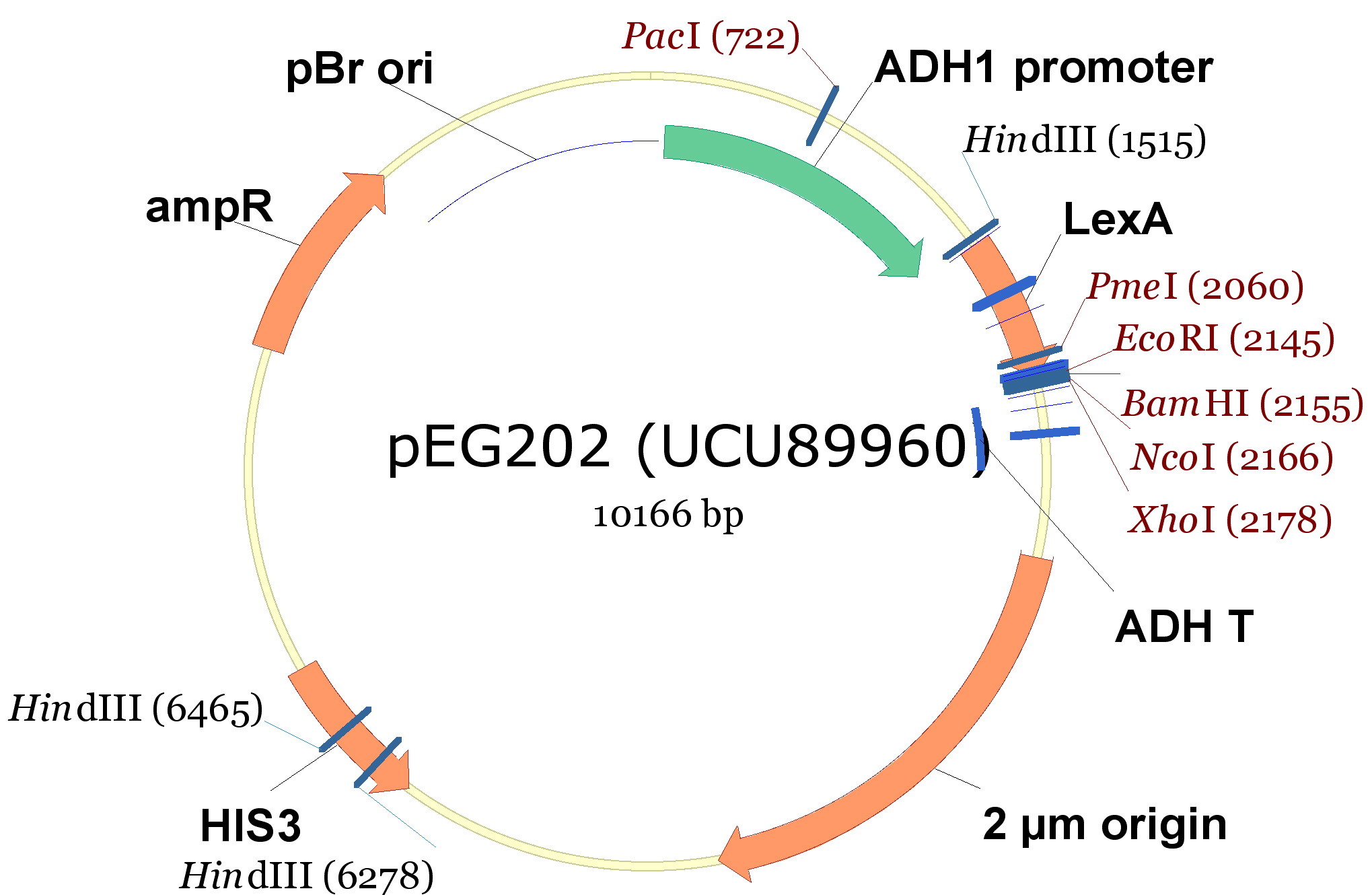
| 340px |
The cloning we did was:
| <partinfo>BBa_K792009</partinfo> (PoliHa) | <partinfo>BBa_K792010</partinfo> (TRPZipper2) | <partinfo>BBa_K792011</partinfo> (PoliHb) | <partinfo>BBa_K792012</partinfo> (PoliWb) | |
|---|---|---|---|---|
| pCM182 (TRPa) | X | X | ||
| pCM185 (TRPb) | X | X | ||
| pEG202 (HIS) | X | X |
Protocol
- Digestion of plasmids and TRP/HIS export device:
- pCM182; pCM 185; BBa_K792010 y BBa_K792012 were digested with BamHI and Pst1 restriction enzymes.
- pEG202; BBa_K792009 y BBa_K792011 were digested with BamHI and Not1.
- Purification of digested vectors (pCM185; pCM182; pEG202)
- Ligation of vectors and devices according the anterior table (T4 ligase protocol, overnight)
- Transformation of E. Coli DH5a with the ligation products. Bacterias were plated on LB-agar with Ampicillin, and incubated over night at 37 °C.
- Colonies were used for liquid cultures (LB + Ampicillin) and minipreps were made.
- Constructions (vector + insert) were checked by digestion with restriction enzymes, and 1%-agarose gel (1kb and 100bp as markers).
Once obtained the desired constructions, we transformed yeast strains:
- TCY3081: W303, bar1-, ura3::PAct1-YFP
- TCY3128: W303, bar1-, leu2:: Pprm1-CFP 405
We got the following transformed strains:
|
|
|
|
Week 3: Synthetic Ecology Characterization
Our task is to characterize our system including the devices functioning. We want specifically to:
- Quantify the export of Trp as proof that the devices work
- Determine experimentally some of the parameters that we used in the model
- Characterize the growth of the transformed strains in coculture
In order to characterize the devices that we used to transform our strains we came up with the series of assays that we describe below.
Devices characterization
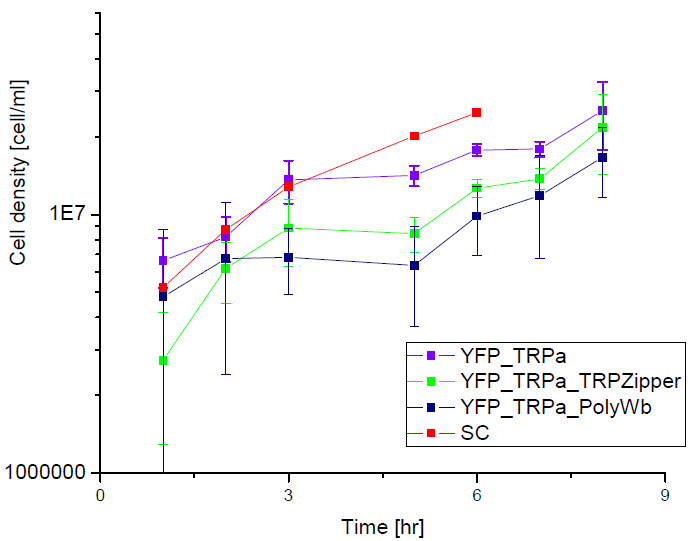
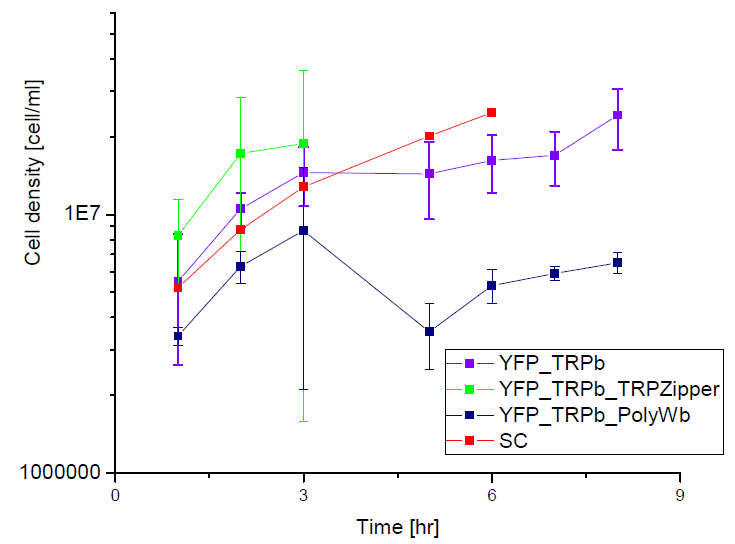
Secretion Rate of Trp as a function of culture growth
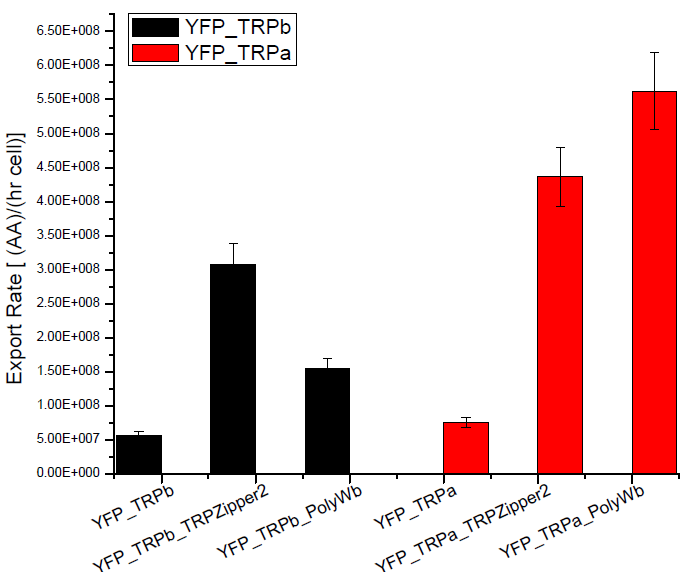
The first step was to actually check if the construct works: do the transformed yeast strains - with the tryptophan devices- actually secrete tryptophan into the medium?
To test this we used the following strains:
|
| ||||||||
|
Protocol
- We started 5ml cultures with 3 replica until they reached exponential phase, overnight, using a -T medium.
- Starting OD for the assay 0.1 (exponential phase).
- We measured OD every hour until they reached an OD: 0.8 (5 hs approximately).
- We measured the Trp signal for each culture medium using the spectrofluorometer.
NOTE: The fluorescence meausurements taken for the Tryptophan in the medium, take into account both the aminoacid secreted by the device and the one diffused.
Therefore the secretion rates calculated will be higher than the actual ones. We used an empty plasmid as control to study tryptophan diffusion.
We used a simple model to measure the secretion rate for all the strains. Since the cultures are in exponential phase, we take
After a few calculations, we find that
Results
Next we show the average OD for each strains used, needed to calculate the secretation rates of Trytophan.
Figure X. check
Figure X. check
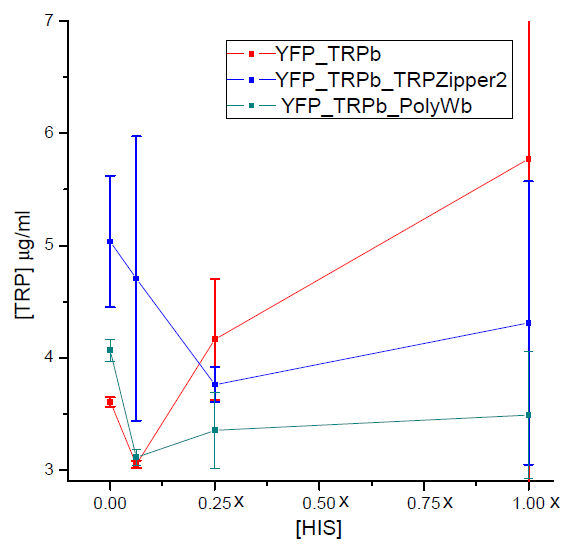
Tryptophan secretion at increasing histidine concentrations
We asked ourselves which was the dependance of tryptophan secretion on the amount of histidine in medium. We carried on this test in order to determine whether the secretion of tryptophan depends on the concentration of another aminoacid in the media, such as histidine and what would be the necessary amount of histidine in medium for the start of our system.
In this experiment we used strains:
| YFP_TRPb_TRPZipper2 | YFP_TRPb_PolyWb | YFP_TRPb (control) |
Protocol
- Starters of each strain used were grown over night in -T media, at 30 °C in shaker.
- After 12 hs, cells were pelleted and washed with -HT media.
- Cultures with increasing concentrations of histidine (0X, 1X, 1/4X and 1/16X) were set at an initial OD: 0.1, for each strain with 2 replica.
- We left the cultures in shaker at 30ºC for 5 hours. After that time, we measured the final OD reached by cultures with the use of a spectrophotometer and the amount of tryptophan present in medium with a spectrofluorometer.
Results
Strain characterization
Experimental determination of strains death rate
We set out to determine how long can auxotroph cells[link] survive in media that lacks both Trytophan and Histidine. These values are the death parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis for simplicity but now we would like to test whether this approximation is accurate.
Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our system's feasability.
Protocol
For this experiment we used
| YFP Strain | CFP Strain |
- We set cultures of the two auxotroph strains without being transformed (YFP and CFP) in medium –HT at an initial OD of 0.01.
- Each day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain.
Result
| Strain | Replica | Monday | Tuesday | Wednesday |
|---|---|---|---|---|
| CFP | 1 | 260 | 320 | 285 |
| CFP | 2 | 267 | 314 | 76 |
| CFP | 3 | 413 | 362 | 278 |
| YFP | 1 | 230 | 316 | 688 |
| YFP | 2 | 291 | 194 | 524 |
| YFP | 3 | 449 | 344 | 725 |
Table: Number of colonies counted per plate.
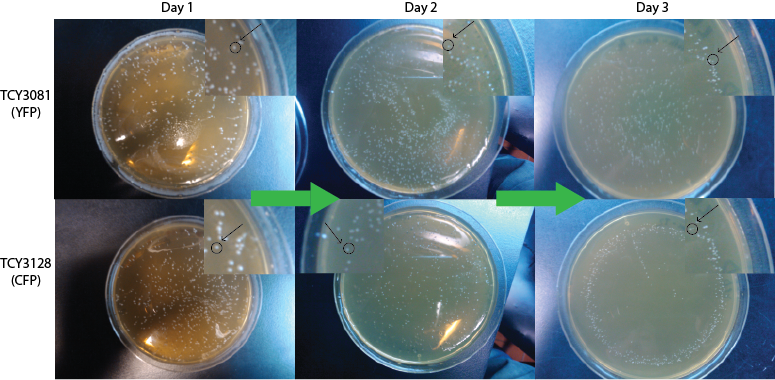
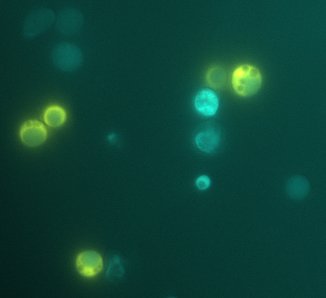
We expect to see a decrease in the number of colonies - because of cell death. We found that this was not the case, in the experiment's time lapse. However we observed that the size of the colonies was smaller everyday as can be seen in the following pictures.
We can infer from this data that though they have not died, they may have enter into a ...Alan state. In this way cells can survive for a period of time in media defficient in amino acid (at least, during the time course of our experiment), but grow slower. Probably this would require more time than 3 days to observe significative cell dying.
| 100px | 100px |
FigureX.
Growth dependence on the Trp and His concentration
An important thing in order to characterize the system is the dependence of the growth rate on the culture with the concentration of the crossfeeding aminoacids, tryptophane (Trp) and histidine (His).
This would allow us to estimate one of the parameters used in our model (the EC50, which is the amount of aminoacid at which the culture reaches half of the maximum growth).
Protocol
1. For this experiment we would use strains YFP and CFP (non-transformed, without devices). We started cultures of 5 ml of initial OD: 0.025 for:
| Strain YFP at the following media [Trp] = 1x; 0.5x; 0.25x; 0.125x; 0.0625x, 0.03125x |
| Strain CFP at the following media [His] = 1x; 0.5x; 0.25x; 0.125x; 0.0625x, 0.03125x |
| Strain YFP at Synthetic Complete media |
| Strain CFP at Synthetic Complete media |
2. Cultures would be left overnight (12 hs) at 30°C with agitation and we measured OD reached with the use of spectrophotometer.
Results
To be done with our strains soon.
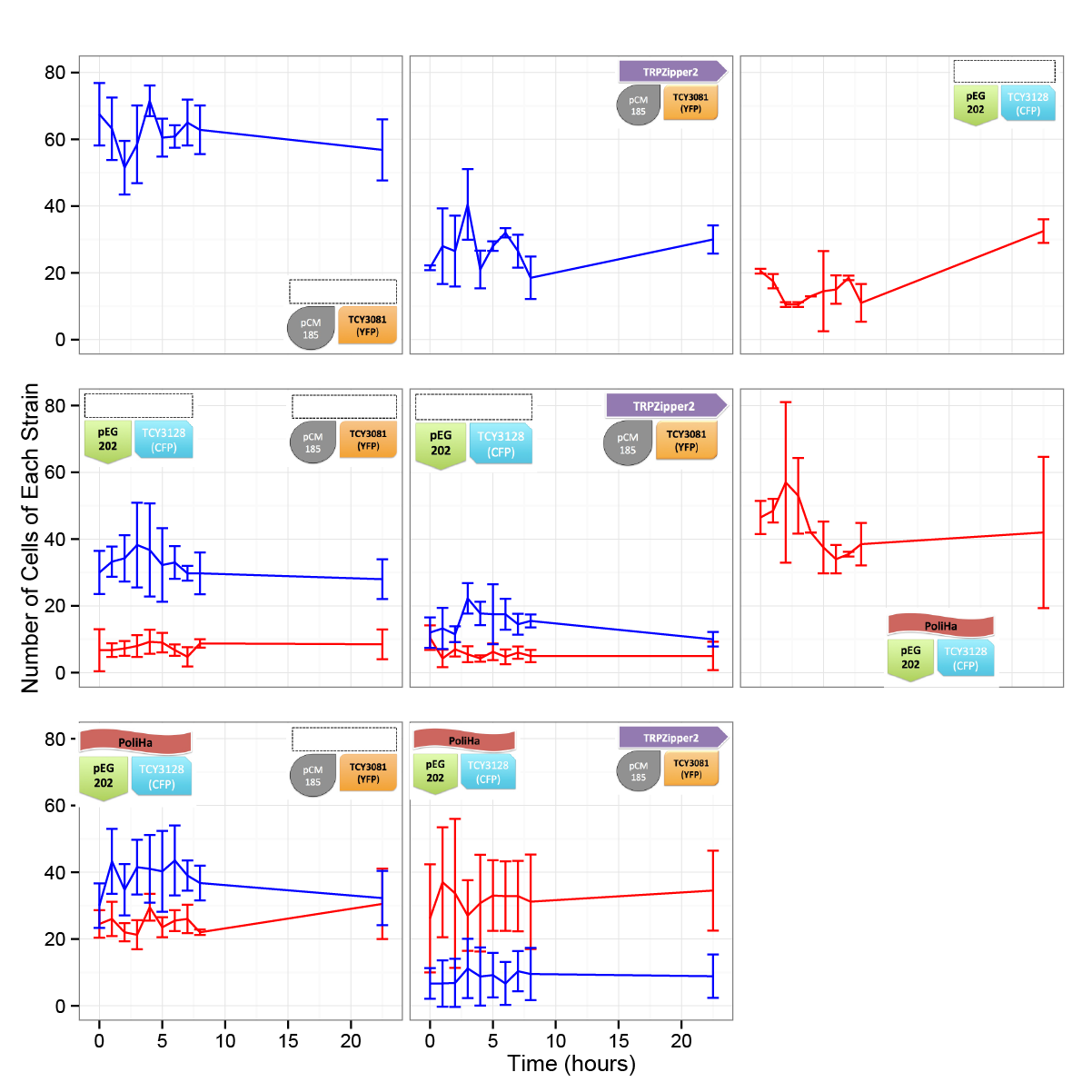
Coculture of transformed strains
We did the first experiment to test whether our system works and how two transformed strains grow together.
We designed an assay to do so; following the growth of these strains:
| CFP_His | YFP_TRPb |
|
Protocol
|
|
|
| 384 wells plate to be used for epifluorescence microscope. |
 "
"