Team:KAIST Korea/Project Background
From 2012.igem.org
(Difference between revisions)
| (30 intermediate revisions not shown) | |||
| Line 39: | Line 39: | ||
{ | { | ||
width:200px; | width:200px; | ||
| - | height:30px; | + | height:30px; |
position:relative; | position:relative; | ||
background:#f2f3f3; | background:#f2f3f3; | ||
padding:5px 30px 5px 30px; | padding:5px 30px 5px 30px; | ||
border:1px solid #f4f4f4; | border:1px solid #f4f4f4; | ||
| - | margin : 0 20px 0 20px; | + | margin : 0 20px 0 20px; |
} | } | ||
| Line 60: | Line 60: | ||
{ | { | ||
font-family: 'Open Sans', sans-serif; | font-family: 'Open Sans', sans-serif; | ||
| - | font-size: | + | font-size:25pt; |
color: #08185d; | color: #08185d; | ||
| + | font-style:italic | ||
padding:0px 0px 0px 10px; | padding:0px 0px 0px 10px; | ||
} | } | ||
| Line 97: | Line 98: | ||
#kaistcontent a { | #kaistcontent a { | ||
| - | + | color:#5a5b5e; | |
} | } | ||
| Line 108: | Line 109: | ||
{ | { | ||
padding: 0px 0px 0px 30px; | padding: 0px 0px 0px 30px; | ||
| + | line-height:140%; | ||
| + | } | ||
| + | #kaistcontent #little2 | ||
| + | { | ||
| + | padding: 0px 0px 0px 15px; | ||
line-height:140%; | line-height:140%; | ||
} | } | ||
| Line 141: | Line 147: | ||
<div id="kaistcontent"> | <div id="kaistcontent"> | ||
| - | + | ||
| - | <div align='center'> | + | <div align='center'> |
| - | <a href="#1st"><span id="tab"> | + | <a href="#1st"><span id="tab"> <b>Overview</b> </span></a> |
| - | <a href="#2nd"><span id="tab"> | + | <a href="#2nd"><span id="tab"><b>Filp Flop</b></span></a> |
| - | </div> | + | <a href="#3rd"><span id="tab"><b>Reguli</b></span></a> |
| - | </br> | + | </div> |
| - | <section id=" | + | </br> |
| + | <section id="1st"> | ||
| + | <div> | ||
| + | <img id="starter-grad" style="height:60px;" src="https://static.igem.org/mediawiki/2012/9/95/Starter_gradient_kaist.png"></img> | ||
| + | <h1>Overview</h1> | ||
| + | |||
| + | </br> | ||
| + | </div> | ||
| + | |||
| + | <img src="https://static.igem.org/mediawiki/2012/e/e1/KAIST_lionLion.png" style="float:left;padding:10px 20px 10px 0px;margin:0px 0px 20px 0px;"/> | ||
| + | </br> | ||
| + | <span id="little"><span id="starter">W</span>hat we see and handle in lab is mostly ‘microscopic’. But there are macroscopic stuffs out there! At night, we can see stars, and among them, we can find Lion’s heart, ‘Regulus’. Although it is not the brightest kind, Regulus still makes it easy to notice the Lion in the sky. </span></br></br> | ||
| + | <span id="little2">Like Regulus, we want our module to be helpful for others to find their own Lion in this universe of synthetic biology. Our module may not be magnificent as that of other teams but we don’t actually want to excel others. We rather want to facilitate the advance of synthetic biology. Like the heart which beats in autonomous manner, our module will run by itself. Regarding all these aspects, we call our project <span style="color:#ff0000;"><b>‘Reguli’</b></span>.</br></br></span> | ||
| + | <div clear='both'></div> | ||
| + | |||
| + | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | ||
| + | |||
| + | <span style="font-size:20px;font-weight:bold">Why does Autoregulation so necessary?</span></br></br> | ||
| + | <span id="little">When we bring external pathway into E.coli to mass-produce the protein, it may collide with E.coli’s innate pathways by consuming critical resources which generate the redox potential (ATP, NADH, or NADPH molecules). This means that E.coli has to reach certain reducing potential to perform exotic metabolic pathway in the cell. </span></br></br> | ||
| + | <span id="little">In conventional methods, the promoters controlling protein expression are repressed until the cells get capacity to produce the protein. Then, this promoter can be induced by putting inducer to the solution on proper cell phase; cell phase can be measured by O.D. value. This kind of method was used because we assume that cell cultural Optical Density (which synchronizes with cell number in the culture) represents inner-cell reduction potential. Although in many time this method successfully makes cells to induce, there are several downsides in this method. </span></br></br> | ||
| + | <span id="little">First of all, problem arises with the process of adding inducers like IPTG or arabinose into media. Because the induction time is decided only by considering cell density (O.D.value) not the capacity of the cells (reduction potential), products may not come out as we predicted. Also, it is an annoying job to measure optical density, finding optimal induction conditions. Even more, if we miss the right time point, we might have to throw them away and start from the first place...</span></br></br> | ||
| + | <span id="little">This is why we came up with our auto-regulative device. As the result of iGEM project this summer, we suggest an auto-regulation module free from induction which utilizes dual-phase switching system.</span></br> | ||
| + | |||
| + | </div> | ||
| + | </br> | ||
| + | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | ||
| + | |||
| + | <span style="font-size:20px;font-weight:bold">Auto-regulative device</span></br></br> | ||
| + | <div align='center'><img src="https://static.igem.org/mediawiki/2012/8/83/KAIST_figure_overview_NUM1.png"/></div></br> | ||
| + | <span id="little">With our automatically-regulative device above, we don’t have to do anything at all to induce gene. When the cells have the capacity to produce the target molecule, the pathway will be turn on. When the cell lost the capacity it will turn off. This is done by cell-source sensor and sensor-generated integrase gene. Integrase and excisionase invert and retrieve the recombination site which exists on the left and right side of promoter. After integrase gene inverts the promoter orientation, genes encoding bio-product pathway are transcribed, making bio-products. In addition, excisionase gene is expressed, reversing promoter orientation to its original state. </span> | ||
| + | </div> | ||
| + | </br> | ||
| + | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | ||
| + | |||
| + | <span style="font-size:20px;font-weight:bold">Environmental Impact</span></br></br> | ||
| + | <div align='center'><img src="https://static.igem.org/mediawiki/2012/c/ce/KAIST_figure_overview_NUM2.png"/></div></br> | ||
| + | <span id="little">Auto-regulating device can be further applied to make a noble sensing/degrading device for toxic compound or pollutant. When cells detect certain amount of toxic or pollutant, sensor induce integrase gene, thus invert the promoter sequence in between two recombination sites. After that, scavenger is generated degrading toxic/pollutant molecule. This novel sensing/scavenging device has its advantage in taking care of the environment and relieving the stress of host cell in automatic manner.</span> | ||
| + | </div> | ||
| + | |||
| + | </br></br> | ||
| + | |||
| + | <div align='right'><a href="#top">▲Back to the top</a></div> | ||
| + | |||
| + | |||
| + | </section> | ||
| + | </br></br> | ||
| + | <section id="2nd"> | ||
<div align='center'> | <div align='center'> | ||
<img src="https://static.igem.org/mediawiki/2012/9/9f/KAIST_Proj_Overview.png"/></br></br> | <img src="https://static.igem.org/mediawiki/2012/9/9f/KAIST_Proj_Overview.png"/></br></br> | ||
</div> | </div> | ||
| - | + | As the name tells us, our new biobrick part <span style="color:#aa0000;"><b>FlipFlop(BBa_K907002 and BBa_K907003)</b> </span> is a noble device which can generate binary signal.</br></br> | |
<span id="little">This part is composed of three sub-parts.</br></br></span> | <span id="little">This part is composed of three sub-parts.</br></br></span> | ||
<div style="border:2px solid;padding:0px 20px 0px 20px;"> | <div style="border:2px solid;padding:0px 20px 0px 20px;"> | ||
| Line 168: | Line 220: | ||
</div> | </div> | ||
</br> | </br> | ||
| - | + | Initially, the FlipFlop promotes the transcription and translation of down-stream gene Protein A according to its promoter orientation. | |
<div align='center'> | <div align='center'> | ||
<br><img style="width:340px;" src="https://static.igem.org/mediawiki/2012/a/ae/KAIST_Proj_Overview3_2.PNG"/> | <br><img style="width:340px;" src="https://static.igem.org/mediawiki/2012/a/ae/KAIST_Proj_Overview3_2.PNG"/> | ||
</div> | </div> | ||
| - | When <a href="http://partsregistry.org/Part:BBa_K907000">BBa_K907000</a>(Mycobacteriophage Bxb1 integrase, Bxb1_Int) recognizes and inverts the sequence flanked | + | When <a href="http://partsregistry.org/Part:BBa_K907000">BBa_K907000</a>(Mycobacteriophage Bxb1 integrase, Bxb1_Int) recognizes and inverts the sequence flanked by attB and attP sequences, promoter orientation is reversed, leaving recombined attL and attR sequences. |
</div> | </div> | ||
</br></br> | </br></br> | ||
| Line 198: | Line 250: | ||
</div> | </div> | ||
| - | </br></br> | + | </br></br> |
| - | <div align='right'><a href="#top">▲Back to the top</a></div> | + | <div align='right'><a href="#top">▲Back to the top</a></div></br> |
</section> | </section> | ||
| - | + | <br> | |
| - | <section id=" | + | <br> |
| - | + | <br> | |
| - | + | </br> | |
| - | + | </br> | |
| + | </br> | ||
| + | <section id="3rd"> | ||
| + | <div> | ||
| + | <img id="starter-grad" style="height:60px;" src="https://static.igem.org/mediawiki/2012/9/95/Starter_gradient_kaist.png"></img> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/9/99/KASIT_Lion2_igem.png" style="float:left;position: relative;z-index: 100;top: -75px;left:5px;"> | ||
| + | <h1>Reguli</h1> | ||
| + | |||
| + | </br> | ||
| + | </div> | ||
| + | |||
<div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | ||
<div align='center'> | <div align='center'> | ||
| Line 213: | Line 275: | ||
</div> | </div> | ||
</br> | </br> | ||
| - | Using the FlipFlop device, we can generate biological machine that | + | Using the FlipFlop device, we can generate biological machine that controls the metabolism as we intended. We decided to call the system as "Auto-regulating system in E.coli", <span style="color:#aa0000;"><b>Reguli</b></span>. To demonstrate the idea, we applied the device to control bio-indigo production, which is catalyzed by bacterial flavin-containing monooxygenase(bFMO).</br> |
</div> | </div> | ||
| - | </br> | + | </br></br> |
<div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> | ||
| - | <span style="font-size: | + | <span style="font-size:20px;font-weight:bold">What is bFMO?</span></br></br> |
<img src="https://static.igem.org/mediawiki/2012/c/c8/KAIST_Overview_2nd_3.png" style="float:right; padding: 10px 10px 10px 10px;"/> | <img src="https://static.igem.org/mediawiki/2012/c/c8/KAIST_Overview_2nd_3.png" style="float:right; padding: 10px 10px 10px 10px;"/> | ||
| - | Bacterial flavin-containing monooxygenase(bFMO) | + | Bacterial flavin-containing monooxygenase(bFMO) converts indole into isatin, which is then sequentially catalyzed into indigoid compounds emitting indigo color. |
| - | </br>Because the enzyme | + | </br>Because the enzyme utilizes the primary metabolite, we can easily notice the enzyme is working well or not. For the following experiments, engineered bFMO gene from Methylophaga sp. Strain SK1 is kindly provided by Duhee Bang from Yonsei University, Republic of Korea. |
<img src="https://static.igem.org/mediawiki/2012/b/b9/KAIST_Overview_2nd_4.png" style="padding: 30px 10px 10px 10px;"/> | <img src="https://static.igem.org/mediawiki/2012/b/b9/KAIST_Overview_2nd_4.png" style="padding: 30px 10px 10px 10px;"/> | ||
| Line 227: | Line 289: | ||
</div> | </div> | ||
| + | |||
</br></br> | </br></br> | ||
| - | <div style="border:2px solid #000000;padding: | + | <div style="border:2px solid #000000;padding:20px 20px 20px 20px;"> |
| - | <img src="https://static.igem.org/mediawiki/2012/ | + | <span style="font-size:20px;font-weight:bold">Working scheme of Our Design</span></br></br> |
| - | + | <img src="https://static.igem.org/mediawiki/2012/e/eb/KAIST_System_1.png" style="padding: 10px 0px 10px 0px;"/> | |
| - | + | <img src="https://static.igem.org/mediawiki/2012/3/3e/KAIST_pAutoSimple_working_scheme.png" style="padding: 10px 0px 10px 0px;"/> | |
| + | <br> | ||
| + | <br> | ||
| + | </br> | ||
| + | </br> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/8/8f/KASIT_System_2.png" style="padding: 10px 0px 10px 0px;"/> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/d/d0/KAIST_pAuto_working_scheme.png" style="padding: 10px 0px 10px 0px;"/> | ||
| + | |||
| - | |||
</div> | </div> | ||
Latest revision as of 03:39, 27 October 2012


2012 KAIST Korea
Mail : kaist.igem.2012@gmail.com
Twitter : twitter.com/KAIST_iGEM_2012
Facebook : www.facebook.com/KAISTiGEM2012

Project : Overview


Overview
 What we see and handle in lab is mostly ‘microscopic’. But there are macroscopic stuffs out there! At night, we can see stars, and among them, we can find Lion’s heart, ‘Regulus’. Although it is not the brightest kind, Regulus still makes it easy to notice the Lion in the sky.
Like Regulus, we want our module to be helpful for others to find their own Lion in this universe of synthetic biology. Our module may not be magnificent as that of other teams but we don’t actually want to excel others. We rather want to facilitate the advance of synthetic biology. Like the heart which beats in autonomous manner, our module will run by itself. Regarding all these aspects, we call our project ‘Reguli’.
What we see and handle in lab is mostly ‘microscopic’. But there are macroscopic stuffs out there! At night, we can see stars, and among them, we can find Lion’s heart, ‘Regulus’. Although it is not the brightest kind, Regulus still makes it easy to notice the Lion in the sky.
Like Regulus, we want our module to be helpful for others to find their own Lion in this universe of synthetic biology. Our module may not be magnificent as that of other teams but we don’t actually want to excel others. We rather want to facilitate the advance of synthetic biology. Like the heart which beats in autonomous manner, our module will run by itself. Regarding all these aspects, we call our project ‘Reguli’.
Why does Autoregulation so necessary?
When we bring external pathway into E.coli to mass-produce the protein, it may collide with E.coli’s innate pathways by consuming critical resources which generate the redox potential (ATP, NADH, or NADPH molecules). This means that E.coli has to reach certain reducing potential to perform exotic metabolic pathway in the cell.
In conventional methods, the promoters controlling protein expression are repressed until the cells get capacity to produce the protein. Then, this promoter can be induced by putting inducer to the solution on proper cell phase; cell phase can be measured by O.D. value. This kind of method was used because we assume that cell cultural Optical Density (which synchronizes with cell number in the culture) represents inner-cell reduction potential. Although in many time this method successfully makes cells to induce, there are several downsides in this method.
First of all, problem arises with the process of adding inducers like IPTG or arabinose into media. Because the induction time is decided only by considering cell density (O.D.value) not the capacity of the cells (reduction potential), products may not come out as we predicted. Also, it is an annoying job to measure optical density, finding optimal induction conditions. Even more, if we miss the right time point, we might have to throw them away and start from the first place...
This is why we came up with our auto-regulative device. As the result of iGEM project this summer, we suggest an auto-regulation module free from induction which utilizes dual-phase switching system.
Auto-regulative device
 With our automatically-regulative device above, we don’t have to do anything at all to induce gene. When the cells have the capacity to produce the target molecule, the pathway will be turn on. When the cell lost the capacity it will turn off. This is done by cell-source sensor and sensor-generated integrase gene. Integrase and excisionase invert and retrieve the recombination site which exists on the left and right side of promoter. After integrase gene inverts the promoter orientation, genes encoding bio-product pathway are transcribed, making bio-products. In addition, excisionase gene is expressed, reversing promoter orientation to its original state.
With our automatically-regulative device above, we don’t have to do anything at all to induce gene. When the cells have the capacity to produce the target molecule, the pathway will be turn on. When the cell lost the capacity it will turn off. This is done by cell-source sensor and sensor-generated integrase gene. Integrase and excisionase invert and retrieve the recombination site which exists on the left and right side of promoter. After integrase gene inverts the promoter orientation, genes encoding bio-product pathway are transcribed, making bio-products. In addition, excisionase gene is expressed, reversing promoter orientation to its original state.

Environmental Impact
 Auto-regulating device can be further applied to make a noble sensing/degrading device for toxic compound or pollutant. When cells detect certain amount of toxic or pollutant, sensor induce integrase gene, thus invert the promoter sequence in between two recombination sites. After that, scavenger is generated degrading toxic/pollutant molecule. This novel sensing/scavenging device has its advantage in taking care of the environment and relieving the stress of host cell in automatic manner.
Auto-regulating device can be further applied to make a noble sensing/degrading device for toxic compound or pollutant. When cells detect certain amount of toxic or pollutant, sensor induce integrase gene, thus invert the promoter sequence in between two recombination sites. After that, scavenger is generated degrading toxic/pollutant molecule. This novel sensing/scavenging device has its advantage in taking care of the environment and relieving the stress of host cell in automatic manner.


- Promoter : BBa_J23119
- RBS : BBa_B0034
- att sites : Recognition site for BBa_K907000(Mycobacteriophage Bxb1 integrase).
Signal 0


Signal 1


Signal 0



Reguli

What is bFMO?
 Bacterial flavin-containing monooxygenase(bFMO) converts indole into isatin, which is then sequentially catalyzed into indigoid compounds emitting indigo color.
Because the enzyme utilizes the primary metabolite, we can easily notice the enzyme is working well or not. For the following experiments, engineered bFMO gene from Methylophaga sp. Strain SK1 is kindly provided by Duhee Bang from Yonsei University, Republic of Korea.
Bacterial flavin-containing monooxygenase(bFMO) converts indole into isatin, which is then sequentially catalyzed into indigoid compounds emitting indigo color.
Because the enzyme utilizes the primary metabolite, we can easily notice the enzyme is working well or not. For the following experiments, engineered bFMO gene from Methylophaga sp. Strain SK1 is kindly provided by Duhee Bang from Yonsei University, Republic of Korea.

 Bacterial flavin-containing monooxygenase(bFMO) converts indole into isatin, which is then sequentially catalyzed into indigoid compounds emitting indigo color.
Because the enzyme utilizes the primary metabolite, we can easily notice the enzyme is working well or not. For the following experiments, engineered bFMO gene from Methylophaga sp. Strain SK1 is kindly provided by Duhee Bang from Yonsei University, Republic of Korea.
Bacterial flavin-containing monooxygenase(bFMO) converts indole into isatin, which is then sequentially catalyzed into indigoid compounds emitting indigo color.
Because the enzyme utilizes the primary metabolite, we can easily notice the enzyme is working well or not. For the following experiments, engineered bFMO gene from Methylophaga sp. Strain SK1 is kindly provided by Duhee Bang from Yonsei University, Republic of Korea.

Working scheme of Our Design


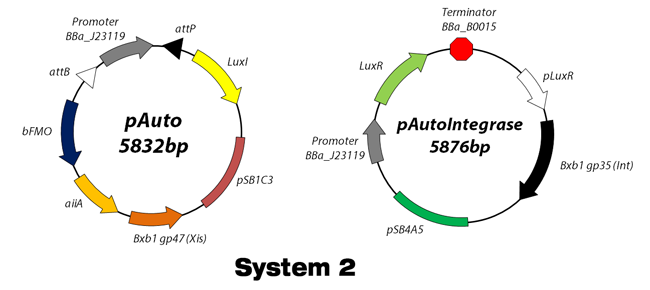





 "
"






