Team:Buenos Aires/Results/Strains
From 2012.igem.org
(→Results) |
(→At different initial OD and proportions) |
||
| (38 intermediate revisions not shown) | |||
| Line 34: | Line 34: | ||
|CFP +(induced YFP) | |CFP +(induced YFP) | ||
|Positive Control | |Positive Control | ||
| + | |- | ||
| + | |TCY 3128 | ||
| + | | style="text-align: center;" |(-H-T) | ||
| + | |CFP | ||
| + | |His device testing | ||
| + | |- | ||
| + | |TCY 3081 | ||
| + | | style="text-align: center;" |(-H-T) | ||
| + | |YFP | ||
| + | |Trp device testing | ||
|} | |} | ||
| rowspan="2" style="text-align: center;" | | | rowspan="2" style="text-align: center;" | | ||
| Line 48: | Line 58: | ||
|} | |} | ||
| - | == | + | == Measurement of strains fluorescence == |
We measured Strains 3281 (YFP) and 3265 (CFP) and got a spectrum of each one prooving that these strains can be distinguished by their fluorescence in culture. | We measured Strains 3281 (YFP) and 3265 (CFP) and got a spectrum of each one prooving that these strains can be distinguished by their fluorescence in culture. | ||
| Line 56: | Line 66: | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File:Bsas2012-strains-Refefluro.png| | + | | align="center" | [[File:Bsas2012-strains-Refefluro.png|300px]] |
|} | |} | ||
| Line 63: | Line 73: | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File: | + | | align="center" | [[File:Fluoro- igembsas2012. strains.png|650px]] |
| - | + | ||
|- | |- | ||
| - | | | + | |'''CFP Fluorescence Screening and YFP Fluorescence Screening''' |
| - | + | ||
|} | |} | ||
| Line 76: | Line 84: | ||
'''Discussion''' | '''Discussion''' | ||
| - | We were able to measure fluorescence in strains 3281 and 3265 using the spectrofluorometer. However, we considered it would not be precise enough for the purposes of measuring cocultures at different proportions. We also noticed a high background noise produced by dead yeast cells at high concentrations, which would make it possible to measure in this way only at a short range of OD while the culture is at exponential phase. | + | We were able to measure fluorescence in strains 3281 and 3265 using the spectrofluorometer. However, we considered it would not be precise enough for the purposes of measuring cocultures at different proportions. We also noticed a high background noise produced by dead yeast cells at high concentrations, which would make it possible to measure in this way only at a short range of OD while the culture is at exponential phase. Therefore we decided to use epifluorescence microscopy (see below). |
| - | == | + | == Strains proportion measurement == |
A more precise way of measuring the proportion of the strains, is with a epifluorescence microscope. | A more precise way of measuring the proportion of the strains, is with a epifluorescence microscope. | ||
| - | We mixed strains 3281 (expresses YFP) and 3265 (expresses CFP) in different proportions and analized the images obtained in the microscope, where we counted cells with different fluorescences. We also did a negative control with a non fluorescent strain (TCY 379). | + | We mixed strains TCY-3281 (that expresses YFP) and TCY-3265 (that expresses CFP) in different proportions and analized the images obtained in the microscope, where we counted cells with different fluorescences. We also did a negative control with a non fluorescent strain (TCY 379). |
'''Description of Mixtures''' | '''Description of Mixtures''' | ||
| - | Mix 1: Negative Control | + | Mix 1: Negative Control Mix 2: 80% CFP; 20%YFP Mix 3: 60% CFP; 40%YFP |
| - | + | ||
| - | Mix 2: 80% CFP; 20%YFP | + | |
| - | + | ||
| - | Mix 3: 60% CFP; 40 | + | |
| - | + | ||
| - | + | ||
| - | Mix 5: 40% CFP; 60%YFP | + | Mix 4: 50% CFP; 50%YFP Mix 5: 40% CFP; 60%YFP Mix 6: 20% CFP; 80%YFP |
| - | |||
'''Results''' | '''Results''' | ||
| Line 103: | Line 104: | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File:Montage-annotated.jpg| | + | | align="center" | [[File:Montage-annotated.jpg|900px]] |
|- | |- | ||
| style="text-align: center;" | Mixtures showing YFP and CFP fluorescence. | | style="text-align: center;" | Mixtures showing YFP and CFP fluorescence. | ||
| Line 114: | Line 115: | ||
'''Counting of cells''' | '''Counting of cells''' | ||
| - | |||
{| class="wikitable" | {| class="wikitable" | ||
| Line 154: | Line 154: | ||
|+ Auxotrophy check | |+ Auxotrophy check | ||
|- | |- | ||
| - | |[[File:Bsas2012-strains-figura3.jpg| | + | |[[File:Bsas2012-strains-figura3.jpg|100px]] |
| - | |[[File:Bsas2012-strains-figura2.png| | + | |[[File:Bsas2012-strains-figura2.png|100px]] |
| + | |[[File:Bsas2012-strains-figura4.png|100px]] | ||
| + | |[[File:Bsas2012-strains-figura5.png|100px]] | ||
|- | |- | ||
| style="text-align: center;" | Medium complete | | style="text-align: center;" | Medium complete | ||
| style="text-align: center;" | Medium without H | | style="text-align: center;" | Medium without H | ||
| + | | style="text-align: center;" | Medium without T | ||
| + | | style="text-align: center;" | Medium without H and T | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
We observed all the strains grew in the SC plate (top left) and only 3154 (+H+T) grew in the -H-T plate (bottom right). In the -T plate (bottom left), only those strains able to synthesize T grew (3265 and 3154) and in the -H plate (top right) only those able to produce H grew (3190 and 3154), as expected. This means our strains work according to their description. We did this several times during the months to check for reversions or contaminations. | We observed all the strains grew in the SC plate (top left) and only 3154 (+H+T) grew in the -H-T plate (bottom right). In the -T plate (bottom left), only those strains able to synthesize T grew (3265 and 3154) and in the -H plate (top right) only those able to produce H grew (3190 and 3154), as expected. This means our strains work according to their description. We did this several times during the months to check for reversions or contaminations. | ||
| Line 269: | Line 265: | ||
{| | {| | ||
|- | |- | ||
| - | |<!--column1-->[[File: | + | |<!--column1-->[[File:HIS-BSAS2012.png|400px]] |
|} | |} | ||
| - | + | As shown in the figure and table there is a basal growth that does not depend on the initial OD or strain proportion. This residual growth produces a growth factor of 6 approximately. | |
| - | + | However we observed a much higher growth at the proportion 1:1 when the initial OD 0.25 and 0.1. This suggests that at these proportions there is a natural cooperation between the strains. The objective of the project is to build upon this natural cooperation and to allow for tunable proportions. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | As shown in | + | |
| - | However we observed a much higher growth at the proportion 1:1 when the initial OD 0.25 and 0.1. | + | |
==== At the same initial OD: 0.2, followed over time ==== | ==== At the same initial OD: 0.2, followed over time ==== | ||
| Line 328: | Line 315: | ||
We repeated this experiment 4 times with different modifications: increasing the amount of days for up to a week, measuring every 12 hs instead of every 24 hs and using different strains. However, bacterial contaminations and the high rate of revertants prevented us from getting to a valid results in those cases, whereas the experiment up to day 2 always worked correctly. This denotes that we should assess the problem of contamination (for example including ampicilin in the cultures) and revertant rate (revising the design of the experiment or looking for more stable strains) as the impossibility to go further than day 2 may put limitations to some applications of the Synthetic Community. | We repeated this experiment 4 times with different modifications: increasing the amount of days for up to a week, measuring every 12 hs instead of every 24 hs and using different strains. However, bacterial contaminations and the high rate of revertants prevented us from getting to a valid results in those cases, whereas the experiment up to day 2 always worked correctly. This denotes that we should assess the problem of contamination (for example including ampicilin in the cultures) and revertant rate (revising the design of the experiment or looking for more stable strains) as the impossibility to go further than day 2 may put limitations to some applications of the Synthetic Community. | ||
| + | |} | ||
== Coculture in Agar and Revertant mutation control == | == Coculture in Agar and Revertant mutation control == | ||
| - | Through this experiment we aim to quantify the rate of revertants of each strain. | + | Through this experiment we aim to quantify the rate of revertants of each strain, and to asses if cross-feeding between a lawn of cells of one strain and colonies from and other strain is posible. |
| - | We used | + | We used petri dishes with agar medium with (+) and without (-) Trp and His as shown in the following table. |
| - | We started a culture of each strain in synthetic complete medium, measured its OD 24 hs after the culture initiated, replaced the synthetic complete medium for one lacking both H and T (to avoid residual growth) and plated 10 6 | + | We started a culture of each strain in synthetic complete medium, measured its OD 24 hs after the culture initiated, replaced the synthetic complete medium for one lacking both H and T (to avoid residual growth) and plated ~10^6 cells (lawn) or ~10^2 cells (seed) as shown by the following table (we considered OD600=1 represents 3*10^7 cells). |
| - | At the same time, 3 controls (one for each strain) were carried in YPD complete medium to check the viability of each strain separately. | + | At the same time, 3 controls (one for each strain) were carried in YPD complete medium to check the viability of each strain separately, and to estimate the seed CFU (colony formin units) more precisely. |
{| class="wikitable" | {| class="wikitable" | ||
! scope="row" style="background: #7ac5e8"|Medium H | ! scope="row" style="background: #7ac5e8"|Medium H | ||
! scope="row" style="background: #7ac5e8"|Medium T | ! scope="row" style="background: #7ac5e8"|Medium T | ||
| - | ! scope="row" style="background: #7ac5e8"| | + | ! scope="row" style="background: #7ac5e8"|Lawn (10^6 cells) |
! scope="row" style="background: #7ac5e8"|Seed (10^2 cells) | ! scope="row" style="background: #7ac5e8"|Seed (10^2 cells) | ||
! scope="row" style="background: #7ac5e8"|Description of experiment | ! scope="row" style="background: #7ac5e8"|Description of experiment | ||
| Line 403: | Line 391: | ||
==== Results ==== | ==== Results ==== | ||
| - | The viability of the strains was high as expected as well as the viability of a control positive strain in the –H-T medium. | + | The viability of the strains was high as expected, as well as the viability of a control positive strain in the –H-T medium. |
| - | As shown in the table, we have a low, but existent, number of revertants from both his and trp auxotrophy strains. This number should be taken into account when interpreting the results from coculture growth after several days, given that the rate of revertants in liquid medium | + | As shown in the table, we have a low, but existent, number of revertants from both his and trp auxotrophy strains. This number should be taken into account when interpreting the results from coculture growth after several days, given that the rate of revertants in liquid medium is probably the same. |
| - | + | ||
| - | == Measurement of Trp in medium | + | Growth in coculture was puzzling, as it resulted in more colonies than the expected. If cooperation was effective, we expected to see as many colonies as "seed" cells, not more. Revertion of cells from the "lawn" doesn't explain the number of colonies either. Probably a combination of both these effects are taking place. |
| + | |||
| + | == Measurement of Trp in medium == | ||
| - | To check the efectiveness of our biobricks, we must first determine the ammount of tryptophan | + | To check the efectiveness of our biobricks, we must first determine the ammount of tryptophan secreted by natural strains to the medium, so we can compare. With that end in mind, we designed a protocol for measurement of tryptophan in medium, based in its fluorescense at 350nm, when excited with 295nm light. |
As a previous step, we checked that none of the other aminoacids used in the medium interferes, by graphically comparing the spectres for uncomplemented medium and medium complemented with leucine, uracile and histidine, at an appropiate range. | As a previous step, we checked that none of the other aminoacids used in the medium interferes, by graphically comparing the spectres for uncomplemented medium and medium complemented with leucine, uracile and histidine, at an appropiate range. | ||
| - | To determine Trp concentration, we must first have a way to transform our readings (intensity) to a more useful output, so we made a calibration curve, through serialized 1:2 dilutions of our medium, which Trp's concentration is | + | To determine Trp concentration, we must first have a way to transform our readings (intensity) to a more useful output, so we made a calibration curve, through serialized 1:2 dilutions of our medium, which Trp's concentration is 20μg/ml, until approximately constant intensity. |
| - | The procedure to measure secretion rates will be growing the strain from a known OD in exponential growth phase in -T medium and plotting it's OD over time, spin-drying at time=t, retrieving the supernatant's Trp concentration and dividing it by the integral of OD vs. time between time=0 and time=t, so we get to a rate which will be proportional to the number of cells in the culture, which means we can actually compare between different strains. Since our medium is free from Trp, all of it should come from within the cells, and if the culture is growing at exponential rates, lysis should be negligible, so the only explanation would be cells exporting their own Trp. | + | The procedure to measure secretion rates will be growing the strain from a known OD in exponential growth phase in -T medium and plotting it's OD over time, spin-drying at time=t, retrieving the supernatant's Trp concentration and dividing it by the integral of OD vs. time between time=0 and time=t, so we get to a rate which will be proportional to the number of cells in the culture, which means we can actually compare between different strains. Since our medium is free from Trp, all of it should come from within the cells, and if the culture is growing at exponential rates, lysis should be negligible, so the only explanation would be cells exporting and diffusing their own Trp. |
{| class="wikitable" border="1" | {| class="wikitable" border="1" | ||
|- | |- | ||
| - | |<!--column1-->[[File: | + | |<!--column1-->[[File:Curva.png | 250px]] |
|- | |- | ||
|Graph:Tryptophan calibration curve | |Graph:Tryptophan calibration curve | ||
| Line 428: | Line 417: | ||
==== Results ==== | ==== Results ==== | ||
| - | + | Through this experiment we can be sure that we would be able to measure increase of Trp in medium as it is exported from the cells, within the biological range of export. | |
| - | The sensitivity of this method seems to be enough to detect concentrations as low as ~0. | + | The sensitivity of this method seems to be enough to detect concentrations as low as ~0.01μg/ml, and as high as 20μg/ml, maybe more. Since our medium is 20μg/ml, we assume that's the saturation point of the curve. If we get bigger intensities than the one corresponding to it, we will dilute the sample. |
| - | + | ||
| - | + | ||
== Growth dependence on the Trp and His concentrations == | == Growth dependence on the Trp and His concentrations == | ||
| Line 447: | Line 434: | ||
! scope="row" style="background: #7ac5e8"|OD Replica 2 | ! scope="row" style="background: #7ac5e8"|OD Replica 2 | ||
|- | |- | ||
| - | |SC | + | |SC (no cells) |
|0,001 | |0,001 | ||
|(-0,0036) | |(-0,0036) | ||
|- | |- | ||
| - | | | + | | -T |
|(-0,003) | |(-0,003) | ||
|(-0,019) | |(-0,019) | ||
|- | |- | ||
| - | | | + | | Trp/2 |
| - | | | + | |2.56 |
| - | | | + | |2.17 |
|- | |- | ||
| - | | | + | | Trp/4 |
| - | | | + | |3.01 |
| - | | | + | |3.11 |
|- | |- | ||
| - | | | + | |Trp/8 |
| - | | | + | |1.54 |
| - | | | + | |1.55 |
|- | |- | ||
| - | | | + | |Trp/16 |
| - | |0 | + | |0.393 |
| - | |0 | + | |0.409 |
|- | |- | ||
| - | | | + | |Trp/32 |
| - | |0 | + | |0.013 |
| - | |0 | + | |0.003 |
|- | |- | ||
| - | | | + | | -H |
|(-0,008) | |(-0,008) | ||
|(-0,012) | |(-0,012) | ||
|- | |- | ||
| - | | | + | | His/2 |
| - | | | + | |3.68 |
| - | | | + | |3.84 |
|- | |- | ||
| - | | | + | | His/4 |
| - | | | + | |2.07 |
| - | | | + | |2.00 |
|- | |- | ||
| - | | | + | |His/8 |
| - | | | + | |1.17 |
| - | |0 | + | |0.97 |
|- | |- | ||
| - | | | + | |His/16 |
| - | |0 | + | |0.47 |
| - | |0 | + | |0.432 |
|- | |- | ||
| - | | | + | |His/32 |
| - | |0 | + | |0.238 |
| - | |0 | + | |0.257 |
|- | |- | ||
| - | | | + | |SC (w/cells) |
| - | | | + | |4.88 |
| - | | | + | |4.91 |
|} | |} | ||
==== Results ==== | ==== Results ==== | ||
| - | As expected the growth has a sigmoidal relationship with the concentration of Trp and His, when plotted in semilogarithmic scale. We call EC50 the effective concentration of each aminoacid at which the culture reaches 50% of the maximal growth. We considered these values as proxies of the Khis and Ktrp parameters of the mathematical model, which can be used to estimate the secretion rate of each aminoacid needed to get effective crossfeeding. | + | As expected the growth has a sigmoidal relationship with the concentration of Trp and His, when plotted in semilogarithmic scale. We call EC50 the effective concentration of each aminoacid at which the culture reaches 50% of the maximal growth. We considered these values as proxies of the Khis and Ktrp parameters of the [[Team:Buenos_Aires/Project/Model#Parameter_selection|mathematical model]], which can be used to estimate the secretion rate of each aminoacid needed to get effective crossfeeding. |
These results can also be observed by comparison of images that show the tubes at different OD. | These results can also be observed by comparison of images that show the tubes at different OD. | ||
| Line 511: | Line 498: | ||
{| class="wikitable" border="1" | {| class="wikitable" border="1" | ||
|- | |- | ||
| - | |<!--column1-->[[File:Bsas2012-strains-alan1.png| | + | |<!--column1-->[[File:Bsas2012-strains-alan1.png|250px]] |
| - | |<!--column2-->[[File:Bsas2012-strains-ultima.jpg| | + | |<!--column2-->[[File:Bsas2012-strains-ultima.jpg|250px]] |
|- | |- | ||
| - | |Images from | + | |Images from HLU series |
| - | |Images from | + | |Images from TLU series |
|} | |} | ||
| Line 521: | Line 508: | ||
SC: Synthetic complete medium with all the aminoacids. It was used as a blank for the spectrofluorometer. | SC: Synthetic complete medium with all the aminoacids. It was used as a blank for the spectrofluorometer. | ||
| - | + | HTLU is the culture in the medium with all the required aminoacids. | |
| + | |||
| + | |||
| + | == Experimental determination of strains death rate== | ||
| + | |||
| + | We set out to determine how long can auxotroph cells[link] survive in media that lacks both Trytophan and Histidine. These values are the '''death''' parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis for simplicity but now we would like to test whether this approximation is accurate. | ||
| + | |||
| + | Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our '' system's feasability''. | ||
| + | |||
| + | ===== Protocol ===== | ||
| + | |||
| + | For this experiment we used | ||
| + | {| | ||
| + | |- | ||
| + | |[[File:BsAs2012-icono-YFP.jpg|200px]] | ||
| + | |[[File:BsAs2012-icono-CFP.jpg|200px]] | ||
| + | |- align="center" | ||
| + | |YFP Strain | ||
| + | |CFP Strain | ||
| + | |} | ||
| + | |||
| + | *We set cultures of the two auxotroph strains without being transformed (YFP and CFP) in medium –HT at an initial OD of 0.01. | ||
| + | *Each day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain. | ||
| + | |||
| + | ===== Result ===== | ||
| + | |||
| + | |||
| + | {| class="wikitable" | ||
| + | ! scope="row" style="background: #7ac5e8" |Strain | ||
| + | ! scope="row" style="background: #7ac5e8" |Replica | ||
| + | ! scope="row" style="background: #7ac5e8" |Monday | ||
| + | ! scope="row" style="background: #7ac5e8" |Tuesday | ||
| + | ! scope="row" style="background: #7ac5e8" |Wednesday | ||
| + | |- | ||
| + | |CFP | ||
| + | |1 | ||
| + | |260 | ||
| + | |320 | ||
| + | |285 | ||
| + | |- | ||
| + | |CFP | ||
| + | |2 | ||
| + | |267 | ||
| + | |314 | ||
| + | |76 | ||
| + | |- | ||
| + | |CFP | ||
| + | |3 | ||
| + | |413 | ||
| + | |362 | ||
| + | |278 | ||
| + | |- | ||
| + | |YFP | ||
| + | |1 | ||
| + | |230 | ||
| + | |316 | ||
| + | |688 | ||
| + | |- | ||
| + | |YFP | ||
| + | |2 | ||
| + | |291 | ||
| + | |194 | ||
| + | |524 | ||
| + | |- | ||
| + | |YFP | ||
| + | |3 | ||
| + | |449 | ||
| + | |344 | ||
| + | |725 | ||
| + | |} | ||
| + | |||
| + | '''Table:''' Number of colonies counted per plate. | ||
| + | |||
| + | We expected to see a decrease in the number of colonies because of cell death. We found that this was not the case in the experiment's time lapse. However we observed that the size of the colonies was smaller everyday as can be seen in the following pictures. | ||
| + | |||
| + | [[File:Bsas2012kdeathcells.png| 500px]] | ||
| + | |||
| - | + | We can infer from this data that though they have not died, they may have enter into a persistant state. In this way cells can survive for a period of time in media defficient in amino acid (at least, during the time course of our experiment), but grow slower. Probably this would require more time than 3 days to observe significative cell dying. | |
| - | + | These results are consistent with the chosen parameters. Moreover, the slower the death rate the bigger the area in the Parameter Space where regulation is feasable. | |
Latest revision as of 04:04, 27 October 2012

Description of strains
Through our experiments we worked with the following strains kindly provided by [http://www.ifibyne.fcen.uba.ar/new/temas-de-investigacion/laboratorio-de-fisiologia-y-biologia-molecular-lfbm/biologia-de-sistemas/dr-alejandro-colman-lerner/ Alejandro Colman-Lerner's] Lab:
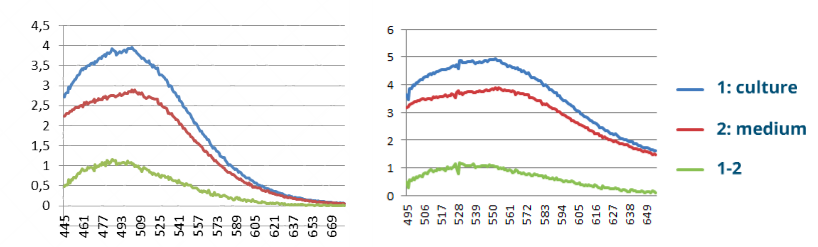
Measurement of strains fluorescence
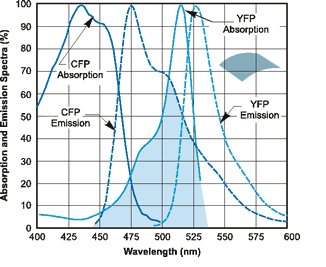
We measured Strains 3281 (YFP) and 3265 (CFP) and got a spectrum of each one prooving that these strains can be distinguished by their fluorescence in culture.
Reference graph Image: YFP and CFP Emission and Absorption Spectra. Obtained from http://flowcyt.salk.edu/fluo.html
|
Results
|
| CFP Fluorescence Screening and YFP Fluorescence Screening |
When measuring YFP Strain 3281, we can see a clear peak around 530 while when measuring CFP Strain 3265, we can see a clear peak around 500, as expected.
Discussion
We were able to measure fluorescence in strains 3281 and 3265 using the spectrofluorometer. However, we considered it would not be precise enough for the purposes of measuring cocultures at different proportions. We also noticed a high background noise produced by dead yeast cells at high concentrations, which would make it possible to measure in this way only at a short range of OD while the culture is at exponential phase. Therefore we decided to use epifluorescence microscopy (see below).
Strains proportion measurement
A more precise way of measuring the proportion of the strains, is with a epifluorescence microscope.
We mixed strains TCY-3281 (that expresses YFP) and TCY-3265 (that expresses CFP) in different proportions and analized the images obtained in the microscope, where we counted cells with different fluorescences. We also did a negative control with a non fluorescent strain (TCY 379).
Description of Mixtures
Mix 1: Negative Control Mix 2: 80% CFP; 20%YFP Mix 3: 60% CFP; 40%YFP
Mix 4: 50% CFP; 50%YFP Mix 5: 40% CFP; 60%YFP Mix 6: 20% CFP; 80%YFP
Results
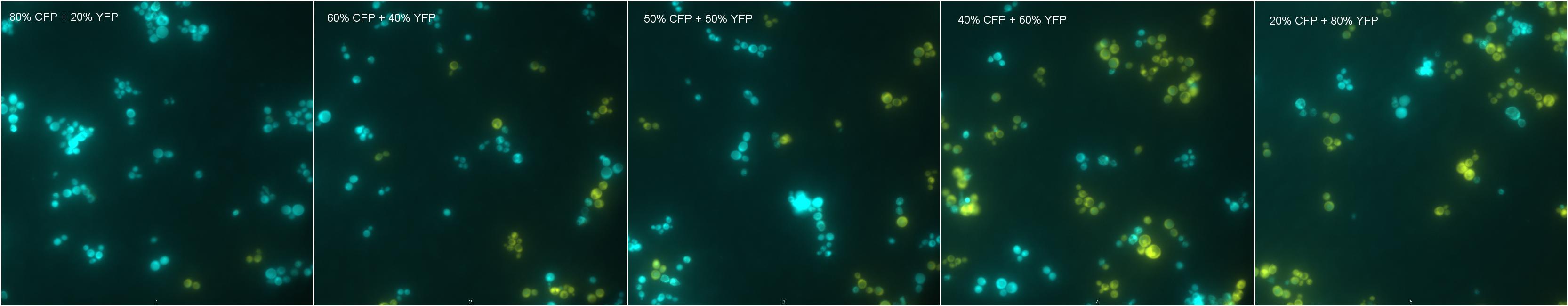
|
| Mixtures showing YFP and CFP fluorescence. |
As shown by images 1-6, cells showing different fluorescences can be count and distinguished from each other in a mixture of strains, and this could be used to measure strains proportion in a coculture.
Counting of cells
| Fluorescence | Mix 1 | Mix 2 | Mix 3 | Mix 4 | Mix 5 | Mix 6 |
| YFP | 0 | 23 | 67 | 115 | 135 | 110 |
| CFP | 0* | 235 | 82 | 107 | 99 | 78 |
The table shows the number of cells counted by expression of fluorescence YFP and CFP in the different mixtures 1-6. I can be observed that the amount of cells is near the proportion stablished by OD measures when preparing the mixtures. This results confirms that epifluorescence measures are reliable and suitable for our research.
Auxotrophy confirmation
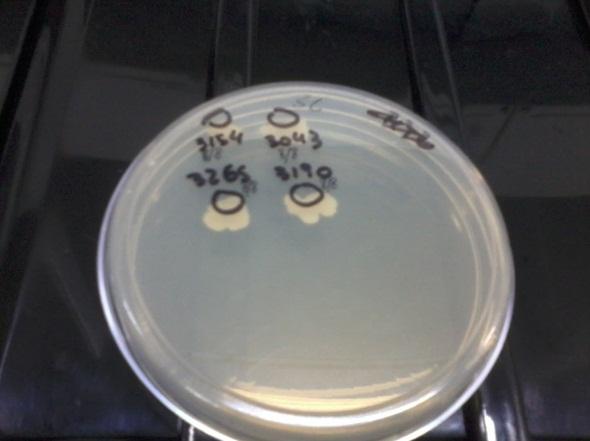
Several times during the experiments we control and checked if the auxotrophies in the selected strain were functional by plating all of them in medium deficient in aminoacids (-H; -T; -H-T and control +H+T). We observed differential growth according to expected due to the description of each strain in point a)
| 
| 
| 
|
| Medium complete | Medium without H | Medium without T | Medium without H and T |
We observed all the strains grew in the SC plate (top left) and only 3154 (+H+T) grew in the -H-T plate (bottom right). In the -T plate (bottom left), only those strains able to synthesize T grew (3265 and 3154) and in the -H plate (top right) only those able to produce H grew (3190 and 3154), as expected. This means our strains work according to their description. We did this several times during the months to check for reversions or contaminations.
Coculture in liquid medium
We used for these experiment TCY3190(H+T-) and TCY3265(H-T+) Positive control: TCY3154 (H+T+) and negative control TCY3043(H-T-)
At different initial OD and proportions
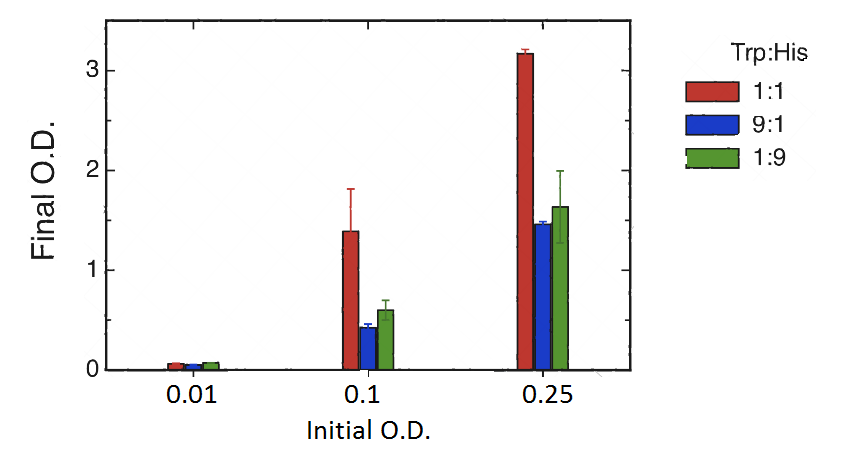
Cultures were set at different initial concentrations (0.25, 0.1 and 0.01) and proportions (1:1; 1:9; 9:1). After 24 hs, we measured OD with the use of a spectrophotometer (two replicas) and we calculated the mean OD and a Growth factor (as Mean OD en time 1 over Initial OD time 0).
| Coculture Proportion (H+T-):(H-T+) | Initial OD(t=0) | OD1 (t=1) | OD2 (t=1) | dilution used for measure t=1 | Mean OD | Growth Factor |
|---|---|---|---|---|---|---|
| 01:01 | 0,25 | 0,32 | 0,314 | 10 | 3,17 | 12,68 |
| 09:01 | 0,25 | 0,148 | 0,144 | 10 | 1,46 | 5,84 |
| 01:09 | 0,25 | 0,138 | 0,189 | 10 | 1,635 | 6,54 |
| 01:01 | 0,1 | 0,109 | 0,169 | 10 | 1,39 | 13,9 |
| 09:01 | 0,1 | 0,04 | 0,045 | 10 | 0,425 | 4,25 |
| 01:09 | 0,1 | 0,067 | 0,053 | 10 | 0,6 | 6 |
| 01:01 | 0,01 | 0,067 | 0,061 | 1 | 0,064 | 6,4 |
| 09:01 | 0,01 | 0,056 | 0,05 | 1 | 0,053 | 5,3 |
| 01:09 | 0,01 | 0,074 | 0,073 | 1 | 0,0735 | 7,35 |
|
As shown in the figure and table there is a basal growth that does not depend on the initial OD or strain proportion. This residual growth produces a growth factor of 6 approximately.
However we observed a much higher growth at the proportion 1:1 when the initial OD 0.25 and 0.1. This suggests that at these proportions there is a natural cooperation between the strains. The objective of the project is to build upon this natural cooperation and to allow for tunable proportions.
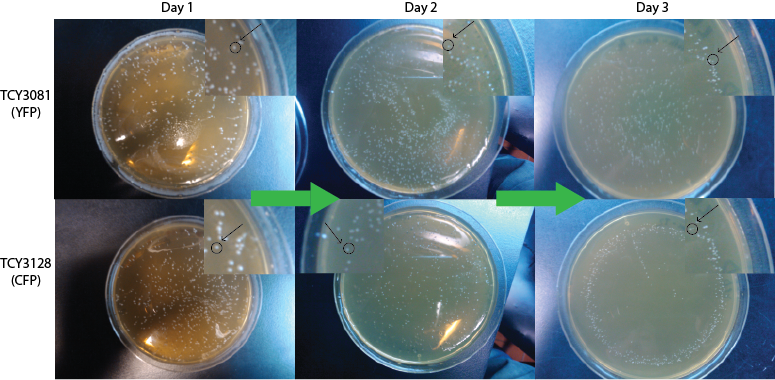
At the same initial OD: 0.2, followed over time
We set the same cultures and cocultures as in point i), but starting all of them at the same OD: 0.2 and we followed them over 2 days. At day 1 we took pictures of them and at day 2 we measured the final OD.
We repeated this experiment 4 times with different modifications: increasing the amount of days for up to a week, measuring every 12 hs instead of every 24 hs and using different strains. However, bacterial contaminations and the high rate of revertants prevented us from getting to a valid results in those cases, whereas the experiment up to day 2 always worked correctly. This denotes that we should assess the problem of contamination (for example including ampicilin in the cultures) and revertant rate (revising the design of the experiment or looking for more stable strains) as the impossibility to go further than day 2 may put limitations to some applications of the Synthetic Community. |
Coculture in Agar and Revertant mutation control
Through this experiment we aim to quantify the rate of revertants of each strain, and to asses if cross-feeding between a lawn of cells of one strain and colonies from and other strain is posible.
We used petri dishes with agar medium with (+) and without (-) Trp and His as shown in the following table.
We started a culture of each strain in synthetic complete medium, measured its OD 24 hs after the culture initiated, replaced the synthetic complete medium for one lacking both H and T (to avoid residual growth) and plated ~10^6 cells (lawn) or ~10^2 cells (seed) as shown by the following table (we considered OD600=1 represents 3*10^7 cells). At the same time, 3 controls (one for each strain) were carried in YPD complete medium to check the viability of each strain separately, and to estimate the seed CFU (colony formin units) more precisely.
| Medium H | Medium T | Lawn (10^6 cells) | Seed (10^2 cells) | Description of experiment | Results after 3 days - Replica 1 | Results after 3 days - Replica 2 |
|---|---|---|---|---|---|---|
| (-) | (+) | (-) | Strain –H+T | Control of His revertants | 7 | 7 |
| (+) | (-) | (-) | Strain +H-T | Control of Trp revertants | 2 | 7 |
| (-) | (-) | Strain +H-T | Strain –H+T | Coculture; we expect to see natural cooperation | 960 | 800 |
| (-) | (-) | Strain –H+T | Strain +H-T | Coculture; we expect to see natural cooperation | 500 | 712 |
| (-) | (-) | (-) | Strain +H+T | Viability of yeasts in medium | 171 | (-) |
Table: Shows description of each plate content and results in number of colonies counted by plate at day 3. YPD control results plates are not shown in the table.
| 
|
| Petri Dishes | With marks of the counting of colonies |
Results
The viability of the strains was high as expected, as well as the viability of a control positive strain in the –H-T medium. As shown in the table, we have a low, but existent, number of revertants from both his and trp auxotrophy strains. This number should be taken into account when interpreting the results from coculture growth after several days, given that the rate of revertants in liquid medium is probably the same.
Growth in coculture was puzzling, as it resulted in more colonies than the expected. If cooperation was effective, we expected to see as many colonies as "seed" cells, not more. Revertion of cells from the "lawn" doesn't explain the number of colonies either. Probably a combination of both these effects are taking place.
Measurement of Trp in medium
To check the efectiveness of our biobricks, we must first determine the ammount of tryptophan secreted by natural strains to the medium, so we can compare. With that end in mind, we designed a protocol for measurement of tryptophan in medium, based in its fluorescense at 350nm, when excited with 295nm light. As a previous step, we checked that none of the other aminoacids used in the medium interferes, by graphically comparing the spectres for uncomplemented medium and medium complemented with leucine, uracile and histidine, at an appropiate range.
To determine Trp concentration, we must first have a way to transform our readings (intensity) to a more useful output, so we made a calibration curve, through serialized 1:2 dilutions of our medium, which Trp's concentration is 20μg/ml, until approximately constant intensity.
The procedure to measure secretion rates will be growing the strain from a known OD in exponential growth phase in -T medium and plotting it's OD over time, spin-drying at time=t, retrieving the supernatant's Trp concentration and dividing it by the integral of OD vs. time between time=0 and time=t, so we get to a rate which will be proportional to the number of cells in the culture, which means we can actually compare between different strains. Since our medium is free from Trp, all of it should come from within the cells, and if the culture is growing at exponential rates, lysis should be negligible, so the only explanation would be cells exporting and diffusing their own Trp.
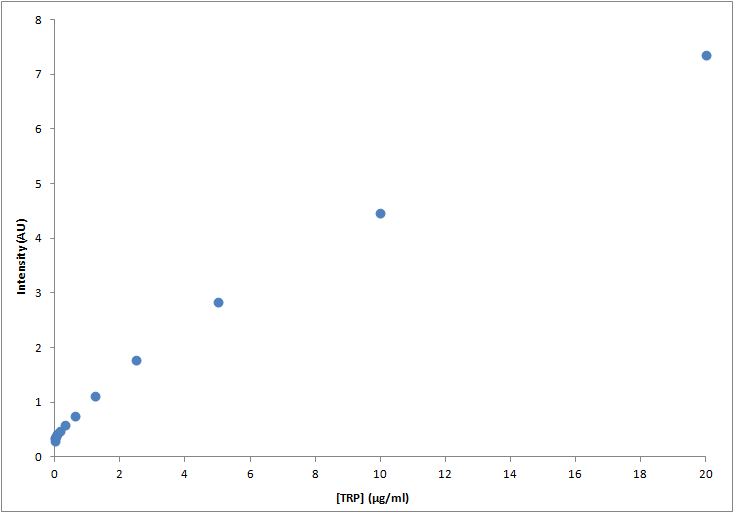
|
| Graph:Tryptophan calibration curve |
Results
Through this experiment we can be sure that we would be able to measure increase of Trp in medium as it is exported from the cells, within the biological range of export. The sensitivity of this method seems to be enough to detect concentrations as low as ~0.01μg/ml, and as high as 20μg/ml, maybe more. Since our medium is 20μg/ml, we assume that's the saturation point of the curve. If we get bigger intensities than the one corresponding to it, we will dilute the sample.
Growth dependence on the Trp and His concentrations
A important thing to characterize of the system is the dependence of the growth rate of the culture with the concentration of the crossfeeding aminoacids, tryptophane (Trp) and histidine (His). To do this we measured the final OD after an overnight growth in medium with different concentrations of Trp and His.
We used strain ACL-379, that is auxotroph for both Trp and His. We prepared serial dilutions of SC medium in –T and –H medium, therefore creating two curves: one with decreasing concentrations of Trp and the other with decreasing concentrations of His. We then inoculated equal amounts of ACL-379 in each tube and incubated them overnight at 30°C with agitation. We took a picture of each tube and measured the OD600 reached by each culture.
| Medium | OD Replica 1 | OD Replica 2 |
|---|---|---|
| SC (no cells) | 0,001 | (-0,0036) |
| -T | (-0,003) | (-0,019) |
| Trp/2 | 2.56 | 2.17 |
| Trp/4 | 3.01 | 3.11 |
| Trp/8 | 1.54 | 1.55 |
| Trp/16 | 0.393 | 0.409 |
| Trp/32 | 0.013 | 0.003 |
| -H | (-0,008) | (-0,012) |
| His/2 | 3.68 | 3.84 |
| His/4 | 2.07 | 2.00 |
| His/8 | 1.17 | 0.97 |
| His/16 | 0.47 | 0.432 |
| His/32 | 0.238 | 0.257 |
| SC (w/cells) | 4.88 | 4.91 |
Results
As expected the growth has a sigmoidal relationship with the concentration of Trp and His, when plotted in semilogarithmic scale. We call EC50 the effective concentration of each aminoacid at which the culture reaches 50% of the maximal growth. We considered these values as proxies of the Khis and Ktrp parameters of the mathematical model, which can be used to estimate the secretion rate of each aminoacid needed to get effective crossfeeding.
These results can also be observed by comparison of images that show the tubes at different OD.

| 
|
| Images from HLU series | Images from TLU series |
Notes: SC: Synthetic complete medium with all the aminoacids. It was used as a blank for the spectrofluorometer.
HTLU is the culture in the medium with all the required aminoacids.
Experimental determination of strains death rate
We set out to determine how long can auxotroph cells[link] survive in media that lacks both Trytophan and Histidine. These values are the death parameters for CFP and YFP strains used in our model[link]. These were taken as equal in the mathematical analysis for simplicity but now we would like to test whether this approximation is accurate.
Given that our system most likely will present a lag phase until a certain amount of both AmioAcids is accumulated in the media, will the cells be viable until this occurs? This is a neccesary check of our system's feasability.
Protocol
For this experiment we used
| YFP Strain | CFP Strain |
- We set cultures of the two auxotroph strains without being transformed (YFP and CFP) in medium –HT at an initial OD of 0.01.
- Each day we plated the same amount of µl of the culture and counted the number of colonies obtain in each plate. We set 3 replica of each strain.
Result
| Strain | Replica | Monday | Tuesday | Wednesday |
|---|---|---|---|---|
| CFP | 1 | 260 | 320 | 285 |
| CFP | 2 | 267 | 314 | 76 |
| CFP | 3 | 413 | 362 | 278 |
| YFP | 1 | 230 | 316 | 688 |
| YFP | 2 | 291 | 194 | 524 |
| YFP | 3 | 449 | 344 | 725 |
Table: Number of colonies counted per plate.
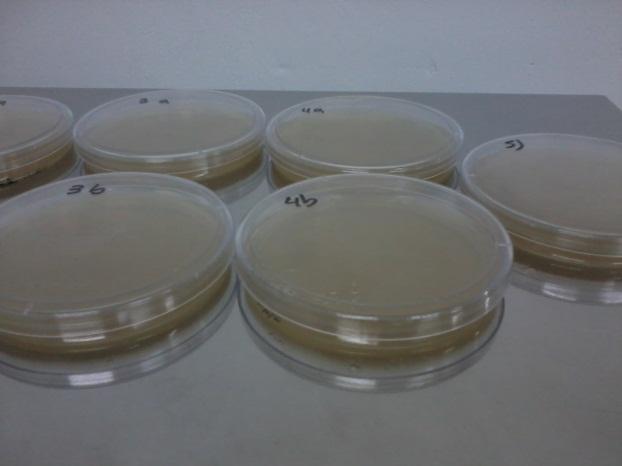
We expected to see a decrease in the number of colonies because of cell death. We found that this was not the case in the experiment's time lapse. However we observed that the size of the colonies was smaller everyday as can be seen in the following pictures.
We can infer from this data that though they have not died, they may have enter into a persistant state. In this way cells can survive for a period of time in media defficient in amino acid (at least, during the time course of our experiment), but grow slower. Probably this would require more time than 3 days to observe significative cell dying.
These results are consistent with the chosen parameters. Moreover, the slower the death rate the bigger the area in the Parameter Space where regulation is feasable.
 "
"