Team:Buenos Aires/Results/Bb1
From 2012.igem.org
Manugimenez (Talk | contribs) (→BioBricks) |
(→References) |
||
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Buenos_Aires/Templates/menu}} | {{:Team:Buenos_Aires/Templates/menu}} | ||
| + | = PoPS -> His/Trp rich peptide export devices = | ||
| - | + | In order for the cross-feeding scheme to work we need each strain to export the amino acid they produce (either Histidine or Tryptophan). To achieve this we created a devices design to secrete to the medium an His (or Trp) rich peptides. | |
| - | + | ||
| - | + | ||
| - | In order for the cross-feeding scheme to work we need each strain to export the amino acid they produce (either Histidine or Tryptophan). To achieve this we created a devices design to secrete to the medium an His (or Trp) rich peptides | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File: | + | | align="center" | [[File:Crossfeeding-device-design_v01.jpg]] |
|- | |- | ||
| - | | align="center" | ''' | + | | align="center" | '''Basic DNA structure of the devices with their constitutive parts''' |
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
* '''Kozak''' consensus sequence for initiation of translation. | * '''Kozak''' consensus sequence for initiation of translation. | ||
* '''Signal peptide''' that targets the product of the gene for secretion. | * '''Signal peptide''' that targets the product of the gene for secretion. | ||
* '''Trojan peptide''' to increase internalization in target cell. | * '''Trojan peptide''' to increase internalization in target cell. | ||
* '''Payload''': this is the exported amino acid rich domain of the protein. | * '''Payload''': this is the exported amino acid rich domain of the protein. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
The input of the devices are ''PoPS'' and the output is secreted amino acids, so the devices are ''PoPS -> exported AA'' transducers. In principle any PoPS generating part can be used. | The input of the devices are ''PoPS'' and the output is secreted amino acids, so the devices are ''PoPS -> exported AA'' transducers. In principle any PoPS generating part can be used. | ||
| Line 120: | Line 77: | ||
== Kozak Sequence == | == Kozak Sequence == | ||
{| | {| | ||
| - | | style="width: | + | | style="width: 75%" | The Kozak sequence is the eukaryotic analog to the bacterial RBS, it is required for efficient initiation of translation. There is only one yeast Kozak sequence in the registry (part [http://partsregistry.org/Part:BBa_J63003 BBa_J63003], distributed in the [http://partsregistry.org/assembly/libraries.cgi?id=42 2012 kit]). Note that this sequence codes for a glutamic acid (E) after the start codon. |
We decided to create a new part (<partinfo>BBa_K792001</partinfo>) using the 5’UTR of the [http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000006108 MFα1] gene of yeast, partly because we used the signal peptide from this gene (see below). This gene is efficiently translated in yeast, and therefore it stands to reason that translation is efficiently initiated on its mRNA. | We decided to create a new part (<partinfo>BBa_K792001</partinfo>) using the 5’UTR of the [http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000006108 MFα1] gene of yeast, partly because we used the signal peptide from this gene (see below). This gene is efficiently translated in yeast, and therefore it stands to reason that translation is efficiently initiated on its mRNA. | ||
| - | | align="center" | | + | | style="width: 25%" align="center" | |
| + | |||
| + | [[File:Bsas2012-Kozak-box.jpg | 200px]] | ||
| + | |||
{| class="wikitable" | {| class="wikitable" | ||
|+ Kozak consensus parts | |+ Kozak consensus parts | ||
| Line 142: | Line 102: | ||
== Signal Peptide == | == Signal Peptide == | ||
| - | The signal peptides target proteins for secretion, effectively exporting the payload. This peptides are cleaved once the protein is in the lumen of the ER, so they won't have any further relevance. | + | {| |
| - | + | | style="width: 75%" | The signal peptides target proteins for secretion, effectively exporting the payload. This peptides are cleaved once the protein is in the lumen of the ER, so they won't have any further relevance. | |
| + | |||
| + | |||
Part <partinfo>BBa_K416003</partinfo> of the registry codes for a yeast signal peptide, based on [Clements 1991]. As an alternative we designed a second part (<partinfo>BBa_K792002</partinfo>) coding for another signal papetide, from the yeast α-mating factor gene ([http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000006108 MFα1]) [Waters et al 1987]. This part is likely to work well when combined with the Kozak sequence from the same gene (<partinfo>BBa_K792001</partinfo>, see above), as the natural 5' end of the MFα1 transcript is reconstituted. Also, because it is a yeast gene, it can be used as is, without any optimization. | Part <partinfo>BBa_K416003</partinfo> of the registry codes for a yeast signal peptide, based on [Clements 1991]. As an alternative we designed a second part (<partinfo>BBa_K792002</partinfo>) coding for another signal papetide, from the yeast α-mating factor gene ([http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000006108 MFα1]) [Waters et al 1987]. This part is likely to work well when combined with the Kozak sequence from the same gene (<partinfo>BBa_K792001</partinfo>, see above), as the natural 5' end of the MFα1 transcript is reconstituted. Also, because it is a yeast gene, it can be used as is, without any optimization. | ||
| - | + | ||
| - | + | | style="width: 25%" align="center" | [[File:Bsas2012-Signal-box.jpg | 200px]] | |
{| class="wikitable" | {| class="wikitable" | ||
| - | |+ | + | |+ Secretion tag parts |
! scope="row" style="background: #7ac5e8" |Part | ! scope="row" style="background: #7ac5e8" |Part | ||
| - | ! scope="row" style="background: #7ac5e8" | | + | ! scope="row" style="background: #7ac5e8" | |
|- | |- | ||
| - | | | + | |<partinfo>BBa_K416003</partinfo> |
| - | | | + | | already in registry |
|- | |- | ||
| - | | | + | |<partinfo>BBa_K792002</partinfo> |
| - | | | + | | our contribution |
| + | |} | ||
| + | |||
|} | |} | ||
== Trojan peptide == | == Trojan peptide == | ||
| - | Trojan peptides are short (15aa) sequences that penetrate through the plasma membrane inside the cell without the need of any receptor or endocitosis process [Derossi 1998]. We want to use them to increase the efficiency with which the payload enters the target cell. Ideally, they should not contain Trp or His, as those are the relevant amino acids for exportation. Two good candidates are the penetratin from the HIV TAT protein, and polyarginine [Jones et al 2005]. | + | {| |
| - | + | |- | |
| + | | Trojan peptides are short (15aa) sequences that penetrate through the plasma membrane inside the cell without the need of any receptor or endocitosis process [Derossi 1998]. We want to use them to increase the efficiency with which the payload enters the target cell. Ideally, they should not contain Trp or His, as those are the relevant amino acids for exportation. Two good candidates are the penetratin from the HIV TAT protein, and polyarginine [Jones et al 2005]. | ||
| + | {| | ||
| + | | | ||
{| class="wikitable" | {| class="wikitable" | ||
|+ Primary protein structure for penetratins | |+ Primary protein structure for penetratins | ||
| Line 177: | Line 144: | ||
|} | |} | ||
| - | This proteins are not from yeast, so we needed to retro-translate them and codon-optimize them for expression in yeast. To do this we used the R package [http://www.bioconductor.org/packages/2.10/bioc/html/GeneGA.html geneGA], that takes into account codon usage and messenger secondary structure in the optimization process. | + | | This proteins are not from yeast, so we needed to retro-translate them and codon-optimize them for expression in yeast. To do this we used the R package [http://www.bioconductor.org/packages/2.10/bioc/html/GeneGA.html geneGA], that takes into account codon usage and messenger secondary structure in the optimization process. |
| + | |} | ||
| + | | style="width: 25%" align="center" | [[File:Bsas2012-Trojan-box.jpg | 200px]] | ||
{| class="wikitable" | {| class="wikitable" | ||
| + | |+ Trojan peptide parts | ||
! scope="row" style="background: #7ac5e8"|Penetratin | ! scope="row" style="background: #7ac5e8"|Penetratin | ||
! scope="row" style="background: #7ac5e8"|Part | ! scope="row" style="background: #7ac5e8"|Part | ||
| - | |||
|- | |- | ||
|TAT | |TAT | ||
|<partinfo>BBa_K792003</partinfo> | |<partinfo>BBa_K792003</partinfo> | ||
| - | |||
|- | |- | ||
|polyarginine | |polyarginine | ||
|<partinfo>BBa_K792004</partinfo> | |<partinfo>BBa_K792004</partinfo> | ||
| - | | | + | |} |
| + | |||
|} | |} | ||
== Payloads == | == Payloads == | ||
| - | The payloads are the elements of the synthetic gene that code for the “amino acid rich” region of the secreted protein. By “a.a. rich” we mean, rich in the amino acid we want to export, Trp or His in our case. These domains should be soluble enough not to cause precipitation of the protein, and should be relatively stable not to be degraded before they are actually secreted from the cell. | + | {| |
| + | |- | ||
| + | |The payloads are the elements of the synthetic gene that code for the “amino acid rich” region of the secreted protein. By “a.a. rich” we mean, rich in the amino acid we want to export, Trp or His in our case. These domains should be soluble enough not to cause precipitation of the protein, and should be relatively stable not to be degraded before they are actually secreted from the cell. | ||
| + | |||
| + | We have used and contributed 4 new payload biobricks. | ||
| + | |||
| + | * '''His-tag''', a polyhistidine-tag normally used for protein purification protocols, or as an epitope for commercial antibodies. | ||
| + | * '''TrpZipper2''', a small peptide that folds into a beta-hairpin secondary structure. The indole rings of the Trp form a hydrophobic core. The protein is water soluble and monomeric [Cochran 2001]. | ||
| + | * '''PolyHb''' and '''PolyWb''', Histidine and Tryptophan rich peptides design by us for this project. | ||
| + | |||
| + | | style="width: 25%" align="center" | [[File:Bsas2012-Payload-box.jpg | 200px ]] | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | |+ Payloads parts | + | |+ Payloads parts |
! scope="row" style="background: #7ac5e8"|Payload | ! scope="row" style="background: #7ac5e8"|Payload | ||
! scope="row" style="background: #7ac5e8"|Part | ! scope="row" style="background: #7ac5e8"|Part | ||
| - | |||
|- | |- | ||
|His tag | |His tag | ||
|<partinfo>BBa_K792005</partinfo> | |<partinfo>BBa_K792005</partinfo> | ||
| - | |||
|- | |- | ||
|TrpZipper2 | |TrpZipper2 | ||
|<partinfo>BBa_K792006</partinfo> | |<partinfo>BBa_K792006</partinfo> | ||
| - | |||
|- | |- | ||
|PolyHb | |PolyHb | ||
|<partinfo>BBa_K792007</partinfo> | |<partinfo>BBa_K792007</partinfo> | ||
| - | |||
|- | |- | ||
|PolyWb | |PolyWb | ||
|<partinfo>BBa_K792008</partinfo> | |<partinfo>BBa_K792008</partinfo> | ||
| - | |||
|} | |} | ||
| - | + | |} | |
| - | + | {| | |
| - | + | |'''PolyHb''' and '''PolyWb''' were designed taking into acount the following consideration: | |
| - | PolyHb and PolyWb were | + | |
# avoided repeating the same residue in tandem to minimize local tRNA depletion | # avoided repeating the same residue in tandem to minimize local tRNA depletion | ||
# avoided tryptophan in tandem because of their low solubility | # avoided tryptophan in tandem because of their low solubility | ||
# we included glycine to avoid the formation of rigid structures | # we included glycine to avoid the formation of rigid structures | ||
# included acidic and basic amino acids to increase solubility | # included acidic and basic amino acids to increase solubility | ||
| - | + | | | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| class="wikitable" | {| class="wikitable" | ||
| - | + | |+ Payloads parts and protein sequences | |
| - | ! scope="row" style="background: #7ac5e8"| | + | ! scope="row" style="background: #7ac5e8"|Payload |
| - | ! scope="row" style="background: #7ac5e8"| | + | ! scope="row" style="background: #7ac5e8"|Residue sequence |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |His tag |
| - | | | + | |HNHNHNHNHNHN |
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | |||
| - | |||
| - | |||
| - | |||
|TrpZipper2 | |TrpZipper2 | ||
| + | |SWTWENGKWTWK | ||
|- | |- | ||
| - | | | + | |PolyHb |
| - | | | + | |HGDHDGHGKHKG HGDHDGHGKHKG HGDHDGHGKHKG |
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | |PolyWb |
| - | | | + | |WGDWDGWGKWKG WGDWDGWGKWKG WGDWDGWGKWKG |
| - | | | + | |} |
| - | + | ||
| - | + | ||
|} | |} | ||
| - | + | Retro-translating and optimizing for yeast (as explained above), we obtained the final sequences (see the registry for more details). | |
| - | + | = Implementation = | |
| - | + | '''Due to time and resources limitation''', we decided to simplify the construction (assembly) process as much as possible. The '''devices were ordered as a whole''' (as gBlocks gene fragments) instead of obtaining each constitutive biobrick part and then assembling them. Although this goes against the standard part base approach, it '''saved both time and money'''. We plan to obtain each constitutive part from the devices by PCR with suffix/prefix containing primers, as a contribution to the registry. | |
| - | + | ||
| - | + | '''We decided to use yeast expression plasmids with repressible or constitutive promoters''', to drive the expression of our devices. This decision was taken because such plasmids were readily available to us, they had adequate selection markers and they are a fairly standard approach in yeast genetics. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File:Bsas2012- | + | | align="center" | [[File:Bsas2012-BBgeneric.jpg|850px]] |
| + | |- | ||
| + | | align="center" | '''Figure 1: DNA structure we sent to synthesis | ||
|} | |} | ||
| + | Although the prefix and suffix are not considered part of the device, they were included in DNA structure we the ordered for easy future assembly. | ||
| + | Because the entire ORF is contained within the prefix and suffix, no care for in-frame assembly has to be taken. We used the original ''RFC10 BioBrick standard''. | ||
| + | In addition, '''we included convenient restriction sites (RS) for directional cloning into the yeast expression vectors''' (''RS1'' and ''RS3''). ''RS2'' will allow easy removal of the sequence coding for the trojan peptide, by restriction and re-ligation. | ||
| - | |||
| - | |||
| - | |||
| + | == Selection of restriction sites == | ||
| - | + | We yet have to choose exactly which restriction enzymes we are going to use, so we can know the final DNA sequence to include in the synthesis request. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | === Considering Yeast Expression Plasmids === | |
| + | {| | ||
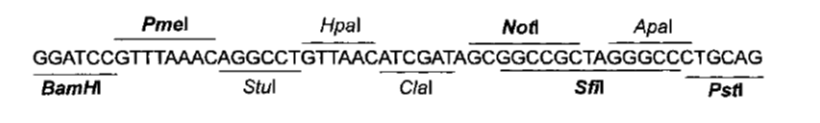
| + | |width = "50%" |To determine which restriction sites to use for cloning (''RS1'' and ''RS3'' in Figure 1), '''we need to know the ''MCS'' of the expression plasmids we are going to use'''. | ||
| - | Comparing Figure 3 and Figure 4, BamHI and NotI appear in both MCS in the same order, so they are good candidates for RS1 and RS3 (Figure 1) respectively. | + | One of these plasmids will probably of the pCM180-5 series, which are centromeric plasmids with TRP1 marker, and with a doxycycline repressible promoter [Gari et al 1996]. |
| + | |||
| + | |||
| + | Comparing Figure 3 and Figure 4, ''BamHI'' and ''NotI'' appear in both ''MCS'' in the same order, so they are good candidates for ''RS1'' and ''RS3'' (Figure 1) respectively. | ||
We will probably need to clone the construct in a general purpose plasmid for manipulation. For instance, to remove the trojan we need to clone the construct into a plasmid, cut it with the RE of RS2, precipitate the DNA (to get rid of the trojan fragment), and religate. For this to work we have to make sure that there is no RS2 in the vector. A common vector for this is pBluescript, wich has the MCS shown in Figure 5. | We will probably need to clone the construct in a general purpose plasmid for manipulation. For instance, to remove the trojan we need to clone the construct into a plasmid, cut it with the RE of RS2, precipitate the DNA (to get rid of the trojan fragment), and religate. For this to work we have to make sure that there is no RS2 in the vector. A common vector for this is pBluescript, wich has the MCS shown in Figure 5. | ||
| + | |||
| + | | | ||
| + | {| style="width: 100%" | ||
| + | |+ '''Figure 3: Multiple Cloning Site (MCS) of the pCM180 series plasmids'''' | ||
| + | | align="center" | [[File:Bsas2012-bb-Fig3.png|450px]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | ---- | ||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File:Bsas2012-bb- | + | |+ '''Figure 4: pEG202 MCS sequence and restriction sites''' |
| + | | align="center" | [[File:Bsas2012-bb-Fig_4.png|450px]] | ||
|} | |} | ||
| + | |} | ||
| + | |||
| + | The other plasmid we might use is ''pEG202'', with a 2μ ori, HIS3 marker and a constitutive promoter (PADH1). | ||
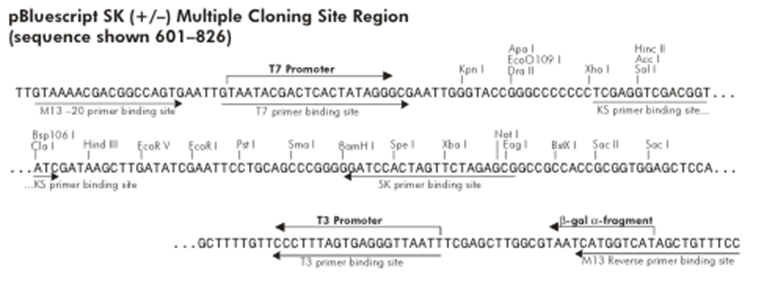
| - | '''Figure 5: pBluescript Multiple Cloning Site''' | + | {| style="width: 100%" |
| + | |+ '''Figure 5: pBluescript Multiple Cloning Site''' | ||
| + | | align="center" | [[File:Bsas2012-bb-Fig5.png|550px]] | ||
| + | |} | ||
| - | == | + | === Considering RFC10 assembly standard restriction enzymes === |
| - | The restriction sites used for RS1-3 have to be different from the ones used in the BioBricks standard. The standard RE for BioBricks are EcoRI, NotI, XbaI, SpeI and PstI. | + | The '''restriction sites used for RS1-3 have to be different from the ones used in the BioBricks standard'''. The standard RE for BioBricks are EcoRI, NotI, XbaI, SpeI and PstI. |
| - | Unfortunately, NotI was our candidate for RS3, so we have a problem here. There are different solutions. We can either use two restriction sites instead of RS3, one for each plasmid, or we can change the BioBrick standard to something like RFC[21] (Berkeley standard) that has no NotI restriction site. | + | Unfortunately, ''NotI'' was our candidate for ''RS3'', so we have a problem here. There are different solutions. We can either use two restriction sites instead of ''RS3'', one for each plasmid, or we can change the BioBrick standard to something like RFC[21] (Berkeley standard) that has no NotI restriction site. |
An other option would be to use the restriction sites in the prefix and suffix to clone the construct into the expression plasmid. This is appealing because we don´t need new REs. Anyhow we would need to include a RS in the 5’ end to be able to directionally clone into pCM180. The new design would look something like Figure 6. | An other option would be to use the restriction sites in the prefix and suffix to clone the construct into the expression plasmid. This is appealing because we don´t need new REs. Anyhow we would need to include a RS in the 5’ end to be able to directionally clone into pCM180. The new design would look something like Figure 6. | ||
| - | |||
| - | |||
{| style="width: 100%" | {| style="width: 100%" | ||
| - | | align="center" | [[File:Bsas2012-bb-Fig6.png| | + | |+ '''Figure 6: Alternative scheme for the restriction sites''' |
| + | | align="center" | [[File:Bsas2012-bb-Fig6.png|850px]] | ||
|} | |} | ||
| - | + | {| | |
| - | + | | If we make RS4 a BamHI site, we can directionally clone the construct into both plasmids (pCM180 and pEG202 ) by cutting with BamHI and NotI. In this design we would not need RS1 and RS3, but we can include them just in case we need to clone them into an other vector. | |
| - | + | ||
| - | If we make RS4 a BamHI site, we can directionally clone the construct into both plasmids (pCM180 and pEG202 ) by cutting with BamHI and NotI. In this design we would not need RS1 and RS3, but we can include them just in case we need to clone them into an other vector. | + | |
Regarding RS2 we need a restriction enzyme that produces cohesive ends, codes for acceptable amino acids, is easily available and not used in an other part of the construct. Some candidate RE are listed in Table 9. | Regarding RS2 we need a restriction enzyme that produces cohesive ends, codes for acceptable amino acids, is easily available and not used in an other part of the construct. Some candidate RE are listed in Table 9. | ||
| - | + | | width = "40%" align="center"| | |
| - | + | ||
{| class="wikitable" | {| class="wikitable" | ||
| + | |+ '''Table 9. Candidate restriction enzymes for RS2''' | ||
! scope="row" style="background: #7ac5e8"|R. Enzyme | ! scope="row" style="background: #7ac5e8"|R. Enzyme | ||
! scope="row" style="background: #7ac5e8"|R. Site sequence | ! scope="row" style="background: #7ac5e8"|R. Site sequence | ||
| Line 349: | Line 317: | ||
|Leu-Glu (LE) | |Leu-Glu (LE) | ||
|} | |} | ||
| - | + | |} | |
| - | + | ||
| - | + | ||
Probably any of them will work, but the trojan peptide needs to be basic so the HindIII site looks better suited. | Probably any of them will work, but the trojan peptide needs to be basic so the HindIII site looks better suited. | ||
| Line 360: | Line 326: | ||
Most likely we wont use RS3, but we can assign it a restriction site just in case. For example NcoI could be used instead of NotI to do the directional cloning into pEG202. | Most likely we wont use RS3, but we can assign it a restriction site just in case. For example NcoI could be used instead of NotI to do the directional cloning into pEG202. | ||
| + | === Final selection === | ||
| + | {| | ||
| + | |- valign="top" | ||
| + | | width = "55%" | | ||
| - | {| class="wikitable" | + | Taking in count everything we mentioned above, this is the final selection restriction enzymes we are going to use. Refer to ''Figure 1'' to check localization of each restriction site. |
| + | | align = "center" | | ||
| + | {| class="wikitable" style="width:75%" | ||
! scope="row" style="background: #7ac5e8"|RS# | ! scope="row" style="background: #7ac5e8"|RS# | ||
! scope="row" style="background: #7ac5e8"|R. Enzyme | ! scope="row" style="background: #7ac5e8"|R. Enzyme | ||
| Line 382: | Line 354: | ||
|G/GATCC | |G/GATCC | ||
|} | |} | ||
| + | |} | ||
| + | |||
| - | + | === References === | |
| - | + | *Clements, J. M., G. H. Catlin, et al. (1991). "Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences." Gene 106(2): 267-271. | |
| + | *Cochran, A. G., N. J. Skelton, et al. (2001). "Tryptophan zippers: stable, monomeric beta -hairpins." Proc Natl Acad Sci U S A 98(10): 5578-5583. | ||
| + | *Derossi, D., G. Chassaing, et al. (1998). "Trojan peptides: the penetratin system for intracellular delivery." Trends Cell Biol 8(2): 84-87. | ||
| + | *Jones, S. W., R. Christison, et al. (2005). "Characterisation of cell-penetrating peptide-mediated peptide delivery." Br J Pharmacol 145(8): 1093-1102. | ||
| + | *Waters, M. G., E. A. Evans, et al. (1988). "Prepro-alpha-factor has a cleavable signal sequence." J Biol Chem 263(13): 6209-6214. | ||
Latest revision as of 02:48, 27 September 2012

Contents |
PoPS -> His/Trp rich peptide export devices
In order for the cross-feeding scheme to work we need each strain to export the amino acid they produce (either Histidine or Tryptophan). To achieve this we created a devices design to secrete to the medium an His (or Trp) rich peptides.

|
| Basic DNA structure of the devices with their constitutive parts |
- Kozak consensus sequence for initiation of translation.
- Signal peptide that targets the product of the gene for secretion.
- Trojan peptide to increase internalization in target cell.
- Payload: this is the exported amino acid rich domain of the protein.
The input of the devices are PoPS and the output is secreted amino acids, so the devices are PoPS -> exported AA transducers. In principle any PoPS generating part can be used.
We built 2 His-export devices, and 2 Trp-export devices:
- His-export I (<partinfo>BBa_K792009</partinfo>)
- Trp export I (<partinfo>BBa_K792010</partinfo>)
- His-export II (<partinfo>BBa_K792011</partinfo>)
- Trp export II (<partinfo>BBa_K792012</partinfo>)
To achive this, we had to create several new basic biobricks:
- Kozak sequence from yeast α-factor mating pheromone (MFα1) (<partinfo>BBa_K792001</partinfo>)
- Secretion tag from yeast α-factor mating pheromone (MFα1) (<partinfo>BBa_K792002</partinfo>)
- HIV TAT penetratin (<partinfo>BBa_K792003</partinfo>)
- Polyarginine trojan peptide (<partinfo>BBa_K792004</partinfo>)
- PolyHa, a Histidine rich peptide (His-Tag) (<partinfo>BBa_K792005</partinfo>)
- TrpZipper2, a Tryptophan rich peptide water soluble and monomeric (<partinfo>BBa_K792006</partinfo>)
- PolyHb, a stable Histidine rich peptide designed by us (<partinfo>BBa_K792007</partinfo>)
- PolyWb, a stable Tryptophan rich peptide designed by us(<partinfo>BBa_K792008</partinfo>)
Details about how we create these new basic biobricks can be found in the next sections. More details can be found in their registry entries also.
The next table summaries each export device composition.
| Device | Kozak | Signal peptide | Trojan peptide | Payload |
|---|---|---|---|---|
| His-export I (<partinfo>BBa_K792009</partinfo>) | MFα1 [-12;6] (<partinfo>BBa_K792001</partinfo>) | MFα1 secretion tag (<partinfo>BBa_K792002</partinfo>) | TAT penetratin (<partinfo>BBa_K792003</partinfo>) | PoliHa (HisTag) (<partinfo>BBa_K792005</partinfo>) |
| Trp-export I (<partinfo>BBa_K792010</partinfo>) | MFα1 [-12;6] (<partinfo>BBa_K792001</partinfo>) | MFα1 secretion tag (<partinfo>BBa_K792002</partinfo>) | TAT penetratin (<partinfo>BBa_K792003</partinfo>) | TrpZipper2 (<partinfo>BBa_K792006</partinfo>) |
| His-export II (<partinfo>BBa_K792011</partinfo>) | <partinfo>BBa_J63003</partinfo> | <partinfo>BBa_K416003</partinfo> | Polyarginine (<partinfo>BBa_K792004</partinfo>) | PolyHb (<partinfo>BBa_K792007</partinfo>) |
| Trp-export II (<partinfo>BBa_K792012</partinfo>) | <partinfo>BBa_J63003</partinfo> | <partinfo>BBa_K416003</partinfo> | Polyarginine (<partinfo>BBa_K792004</partinfo>) | PolyWb (<partinfo>BBa_K792008</partinfo>) |
Kozak Sequence
| The Kozak sequence is the eukaryotic analog to the bacterial RBS, it is required for efficient initiation of translation. There is only one yeast Kozak sequence in the registry (part [http://partsregistry.org/Part:BBa_J63003 BBa_J63003], distributed in the [http://partsregistry.org/assembly/libraries.cgi?id=42 2012 kit]). Note that this sequence codes for a glutamic acid (E) after the start codon.
We decided to create a new part (<partinfo>BBa_K792001</partinfo>) using the 5’UTR of the [http://www.yeastgenome.org/cgi-bin/locus.fpl?dbid=S000006108 MFα1] gene of yeast, partly because we used the signal peptide from this gene (see below). This gene is efficiently translated in yeast, and therefore it stands to reason that translation is efficiently initiated on its mRNA. |
|
Signal Peptide
Trojan peptide
Trojan peptides are short (15aa) sequences that penetrate through the plasma membrane inside the cell without the need of any receptor or endocitosis process [Derossi 1998]. We want to use them to increase the efficiency with which the payload enters the target cell. Ideally, they should not contain Trp or His, as those are the relevant amino acids for exportation. Two good candidates are the penetratin from the HIV TAT protein, and polyarginine [Jones et al 2005].
| 
|
Payloads
| The payloads are the elements of the synthetic gene that code for the “amino acid rich” region of the secreted protein. By “a.a. rich” we mean, rich in the amino acid we want to export, Trp or His in our case. These domains should be soluble enough not to cause precipitation of the protein, and should be relatively stable not to be degraded before they are actually secreted from the cell.
| 
|
PolyHb and PolyWb were designed taking into acount the following consideration:
|
|
Retro-translating and optimizing for yeast (as explained above), we obtained the final sequences (see the registry for more details).
Implementation
Due to time and resources limitation, we decided to simplify the construction (assembly) process as much as possible. The devices were ordered as a whole (as gBlocks gene fragments) instead of obtaining each constitutive biobrick part and then assembling them. Although this goes against the standard part base approach, it saved both time and money. We plan to obtain each constitutive part from the devices by PCR with suffix/prefix containing primers, as a contribution to the registry.
We decided to use yeast expression plasmids with repressible or constitutive promoters, to drive the expression of our devices. This decision was taken because such plasmids were readily available to us, they had adequate selection markers and they are a fairly standard approach in yeast genetics.
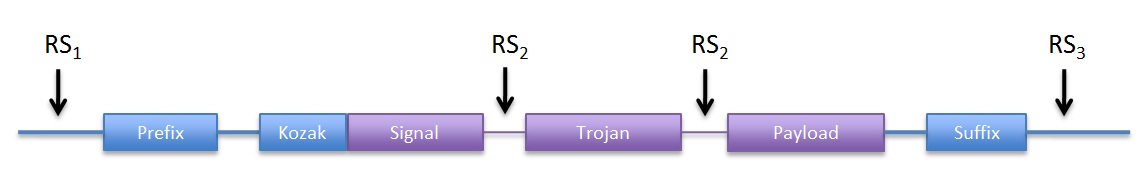
|
| Figure 1: DNA structure we sent to synthesis |
Although the prefix and suffix are not considered part of the device, they were included in DNA structure we the ordered for easy future assembly. Because the entire ORF is contained within the prefix and suffix, no care for in-frame assembly has to be taken. We used the original RFC10 BioBrick standard.
In addition, we included convenient restriction sites (RS) for directional cloning into the yeast expression vectors (RS1 and RS3). RS2 will allow easy removal of the sequence coding for the trojan peptide, by restriction and re-ligation.
Selection of restriction sites
We yet have to choose exactly which restriction enzymes we are going to use, so we can know the final DNA sequence to include in the synthesis request.
Considering Yeast Expression Plasmids
| To determine which restriction sites to use for cloning (RS1 and RS3 in Figure 1), we need to know the MCS of the expression plasmids we are going to use.
One of these plasmids will probably of the pCM180-5 series, which are centromeric plasmids with TRP1 marker, and with a doxycycline repressible promoter [Gari et al 1996].
|
|
The other plasmid we might use is pEG202, with a 2μ ori, HIS3 marker and a constitutive promoter (PADH1).
|
Considering RFC10 assembly standard restriction enzymes
The restriction sites used for RS1-3 have to be different from the ones used in the BioBricks standard. The standard RE for BioBricks are EcoRI, NotI, XbaI, SpeI and PstI.
Unfortunately, NotI was our candidate for RS3, so we have a problem here. There are different solutions. We can either use two restriction sites instead of RS3, one for each plasmid, or we can change the BioBrick standard to something like RFC[21] (Berkeley standard) that has no NotI restriction site.
An other option would be to use the restriction sites in the prefix and suffix to clone the construct into the expression plasmid. This is appealing because we don´t need new REs. Anyhow we would need to include a RS in the 5’ end to be able to directionally clone into pCM180. The new design would look something like Figure 6.
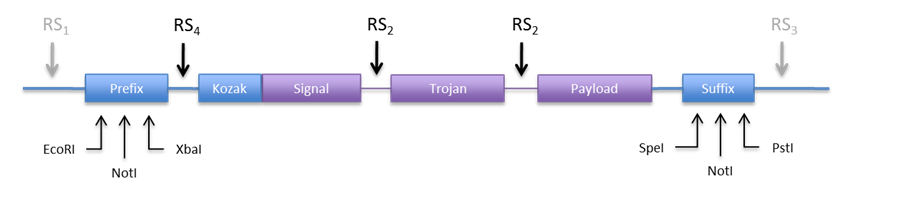
|
| If we make RS4 a BamHI site, we can directionally clone the construct into both plasmids (pCM180 and pEG202 ) by cutting with BamHI and NotI. In this design we would not need RS1 and RS3, but we can include them just in case we need to clone them into an other vector.
Regarding RS2 we need a restriction enzyme that produces cohesive ends, codes for acceptable amino acids, is easily available and not used in an other part of the construct. Some candidate RE are listed in Table 9. |
|
Probably any of them will work, but the trojan peptide needs to be basic so the HindIII site looks better suited. If we want to remove the trojan, we will have to clone the construct into a vector with no HindIII site. One way to do this is to clone it into pBluescript in such a way that the HindIII restriction site of the MCS is removed.
Looking at Figure 5 we can see that if we cut pBluescript with XhoI and PstI, the HindIII site is removed. If we make RS1 -> XhoI (which is easily available) we can cut the construct with these same enzymes and directionally clone it into pBluescript.
Most likely we wont use RS3, but we can assign it a restriction site just in case. For example NcoI could be used instead of NotI to do the directional cloning into pEG202.
Final selection
|
Taking in count everything we mentioned above, this is the final selection restriction enzymes we are going to use. Refer to Figure 1 to check localization of each restriction site. |
|
References
- Clements, J. M., G. H. Catlin, et al. (1991). "Secretion of human epidermal growth factor from Saccharomyces cerevisiae using synthetic leader sequences." Gene 106(2): 267-271.
- Cochran, A. G., N. J. Skelton, et al. (2001). "Tryptophan zippers: stable, monomeric beta -hairpins." Proc Natl Acad Sci U S A 98(10): 5578-5583.
- Derossi, D., G. Chassaing, et al. (1998). "Trojan peptides: the penetratin system for intracellular delivery." Trends Cell Biol 8(2): 84-87.
- Jones, S. W., R. Christison, et al. (2005). "Characterisation of cell-penetrating peptide-mediated peptide delivery." Br J Pharmacol 145(8): 1093-1102.
- Waters, M. G., E. A. Evans, et al. (1988). "Prepro-alpha-factor has a cleavable signal sequence." J Biol Chem 263(13): 6209-6214.
 "
"