Team:USTC-China/background
From 2012.igem.org
MelodyChen (Talk | contribs) |
|||
| (37 intermediate revisions not shown) | |||
| Line 19: | Line 19: | ||
}); | }); | ||
</script> | </script> | ||
| - | |||
| - | |||
<!--this is the top menu--> | <!--this is the top menu--> | ||
| Line 90: | Line 88: | ||
<ol> | <ol> | ||
<li><a href="#2.1"> Genome structure of the toggle switch in lambda phage</a></li> | <li><a href="#2.1"> Genome structure of the toggle switch in lambda phage</a></li> | ||
| - | <li><a href="#2.2"> Function overview of the proteins | + | <li><a href="#2.2"> Function overview of the proteins involved in the toggle switch</a></li> |
<li><a href="#2.3"> Maintenance of lysogeny</a></li> | <li><a href="#2.3"> Maintenance of lysogeny</a></li> | ||
<li><a href="#2.4"> Transformation into Lytic life cycle</a></li> | <li><a href="#2.4"> Transformation into Lytic life cycle</a></li> | ||
| - | <li><a href="#2. | + | <li><a href="#2.5"> Lytic or lysogenic?</a></li> |
</ol> | </ol> | ||
</ul></div></div> | </ul></div></div> | ||
| Line 108: | Line 106: | ||
<a href="https://static.igem.org/mediawiki/2012/5/5a/320px-Tevenphage.png"><img src="https://static.igem.org/mediawiki/2012/5/5a/320px-Tevenphage.png" alt="project image" style="clear:both;width:320;height:229px;"></a><br><small align="center">Diagram of a typical tailed bacteriophage structure</small> | <a href="https://static.igem.org/mediawiki/2012/5/5a/320px-Tevenphage.png"><img src="https://static.igem.org/mediawiki/2012/5/5a/320px-Tevenphage.png" alt="project image" style="clear:both;width:320;height:229px;"></a><br><small align="center">Diagram of a typical tailed bacteriophage structure</small> | ||
</div> | </div> | ||
| - | <p>The phage is a kind of virus that | + | <p>The phage is a kind of virus that infects bacteria. Nowadays, nearly each kind of prokaryote has been found to have its specific phage. Generally, a phage contains a head, a collar and a tail. The head, namely a protein capsid, contains the phage's genetic material and the tail functions as an anchor for binding to the membrane of the bacteria. The phage is tiny and can only be seen under the EMS (electron microscope). The phage has intense infectivity. What's worse, the phage's resistance against the physical and chemical factors is far too stronger than that of bacteria. It can easily survive the toxic organic solvent such as diethyl ether, the chloroform and the severe circumstance such as low temperature and freeze. However, the phage is sensitive to ultraviolet rays and heat.</p> |
| - | <p>The phage widely | + | <p>The phage widely distributes in the nature, especially in the air around the manufacturing shops of the fermentation factory and the drainage system. The phage can spread by air and sneak into nearly every procedure of the fermentation. Main reasons for the pollution of one single fermentation tank include: remaining blind sides both in the tank and its assistant pipes and the misoperation during inoculating. Main sources of the mass invasion of phages include air filtration system, importation of new strains, lysogen and unthoroughly autoclaved media. All these reasons and sources can lead to continuous pollution among tanks and massive infection.</p> |
| - | <p>In fermentation production, the loss results from the | + | <p>In fermentation production, the loss that results from the infection by the phage is striking. Once the bacteria are infected, it is very difficult to eradicate the phages and the lysogens thoroughly because of the intense infection and rapid spread. For this reason, it is quite important to prevent the invasion of the phages. If large tanks of fermentation broth are only polluted slightly, the period of the fermentation will be extended, the broth will become clear gradually and the synthesis of the fermented products will be much more difficult. However, if the fermentation broth is unluckily polluted severely, the production has to stop totally, and massive loss in profit happens afterward. This kind of catastrophe occurs commonly in the fermentation of glutamic acid, amylase, protease, acetone and many kinds of antibiotics. For example, in the fermentation of lactic acid, the production of the starter culture of lactobacillus will stop in 30 minutes once the bacteria are infected severely by phages.</p> |
<p>Some common phenomena that will occur if the fermentation broth is polluted by the phages are listed below:</p> | <p>Some common phenomena that will occur if the fermentation broth is polluted by the phages are listed below:</p> | ||
<li>1. The bacteria will die and lyse in a short time and only the fragments of bacteria will remain afterward.</li> | <li>1. The bacteria will die and lyse in a short time and only the fragments of bacteria will remain afterward.</li> | ||
<li>2. OD at 600nm of the fermentation broth can be found quite low.</li> | <li>2. OD at 600nm of the fermentation broth can be found quite low.</li> | ||
| - | <li>3. Under the microscope, you can see the number of the bacteria is extremely decreased and the bacteria in a colony are abnormally arranged. What's worse, | + | <li>3. Under the microscope, you can see the number of the bacteria is extremely decreased and the bacteria in a colony are abnormally arranged. What's worse, no intact bacteria can be found.</li> |
<li>4. The physicochemical properties are changing. The pH is increasing, the temperature stop climbing and start to decrease gradually. The amount of CO2 emitted is dropping rapidly.</li> | <li>4. The physicochemical properties are changing. The pH is increasing, the temperature stop climbing and start to decrease gradually. The amount of CO2 emitted is dropping rapidly.</li> | ||
<li>5. The metabolism of the bacteria becomes abnormal and the production of the fermentation products stops.</li> | <li>5. The metabolism of the bacteria becomes abnormal and the production of the fermentation products stops.</li> | ||
| Line 132: | Line 130: | ||
<a href="https://static.igem.org/mediawiki/2012/2/25/593px-LambdaPlaques.jpg"><img src="https://static.igem.org/mediawiki/2012/2/25/593px-LambdaPlaques.jpg" alt="project image" style="clear:both;width:200px;height:200px;"></a><br><small align="center">Lysis plaques of lambda phage on E. coli bacteria lambda phage</small> | <a href="https://static.igem.org/mediawiki/2012/2/25/593px-LambdaPlaques.jpg"><img src="https://static.igem.org/mediawiki/2012/2/25/593px-LambdaPlaques.jpg" alt="project image" style="clear:both;width:200px;height:200px;"></a><br><small align="center">Lysis plaques of lambda phage on E. coli bacteria lambda phage</small> | ||
</div> | </div> | ||
| - | <p>Usually, a <a href="">"lytic cycle"</a> ensues, where the lambda DNA is replicated many times and the genes for head, tail and lysis proteins are expressed. This leads to assembly of multiple new phage particles within the cell and subsequent cell lysis, releasing the cell contents, including virions that have been assembled, into the environment</p> | + | <p>Usually, a <a href="https://2012.igem.org/Team:USTC-China/background#2.5">"lytic cycle"</a> ensues, where the lambda DNA is replicated many times and the genes for head, tail and lysis proteins are expressed. This leads to assembly of multiple new phage particles within the cell and subsequent cell lysis, releasing the cell contents, including virions that have been assembled, into the environment</p> |
| - | <p>However, under certain conditions the phage DNA may integrate itself into the host cell chromosome in the <a href="">lysogenic pathway</a>. In this state, the λ DNA is called a prophage and stays resident within the host's genome without apparent harm to the host. The host can be termed a lysogen when a prophage is present.</p><br> | + | <p>However, under certain conditions the phage DNA may integrate itself into the host cell chromosome in the <a href="https://2012.igem.org/Team:USTC-China/background#2.3">lysogenic pathway</a>. In this state, the λ DNA is called a prophage and stays resident within the host's genome without apparent harm to the host. The host can be termed a lysogen when a prophage is present.</p><br> |
<h3><a name="2.1">Genome structure of the toggle switch in lambda phage</a></h3> | <h3><a name="2.1">Genome structure of the toggle switch in lambda phage</a></h3> | ||
| - | <div class="imgholder1" align=" | + | <div class="imgholder1" align="left" style="clear:both;width:289px;height:auto;margin-left:100px;"> |
<a href="https://static.igem.org/mediawiki/2012/6/6f/Bacteriophage_lambda_genome_total.jpg"><img src="https://static.igem.org/mediawiki/2012/6/6f/Bacteriophage_lambda_genome_total.jpg" alt="Bacteriophage_lambda_genome_total" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Schematic representation of the genome of the bacteriophage lambda.</small> | <a href="https://static.igem.org/mediawiki/2012/6/6f/Bacteriophage_lambda_genome_total.jpg"><img src="https://static.igem.org/mediawiki/2012/6/6f/Bacteriophage_lambda_genome_total.jpg" alt="Bacteriophage_lambda_genome_total" style="clear:both;width:289px;height:254px;"></a><br><small align="center">Schematic representation of the genome of the bacteriophage lambda.</small> | ||
</div> | </div> | ||
<div class="imgholder1" align="left" style="float:right;width:235px;height:auto;"> | <div class="imgholder1" align="left" style="float:right;width:235px;height:auto;"> | ||
| - | <a href="https://static.igem.org/mediawiki/2012/ | + | <a href="https://static.igem.org/mediawiki/2012/e/ea/PRM%28new%29.png"><img src="https://static.igem.org/mediawiki/2012/e/ea/PRM%28new%29.png" alt="project image" style="clear:both;width:235px;height:109px;"></a><br><small align="center">Sequence_of_toggle_switch</small> |
</div> | </div> | ||
| - | <p><b>Promoter pR:</b | + | <p><b>Promoter pR:</b>Initializes the rightward transcription. If the pR is not repressed, these proteins will be expressed and the phage will transfer into lytic cycle and plenty of new phage particles will be assembled.</p> |
| - | <p><b>Promoter pRM:</b>Initializes the transcription of repressor CI. It transcripts leftwards. | + | <p><b>Promoter pRM:</b>Initializes the transcription of repressor CI. It transcripts leftwards. The transcription efficiency of pRM is quite low, nearly one eighth of the efficiency of pR.<br> |
| - | OR1, OR2, OR3: Three protein binding regions on the genome of lambda phage. They can be bound by protein CI dimmers(CI2) and Cro | + | <p><b>OR1, OR2, OR3: </b>Three protein binding regions on the genome of lambda phage. They can be bound by protein CI dimmers(CI2) and Cro diamers(Cro2). The sites of these three regions on DNA are shown in the picture below.</p> |
<br> | <br> | ||
<h3><a name="2.2">Function overview of the proteins involve in the toggle switch</a></h3> | <h3><a name="2.2">Function overview of the proteins involve in the toggle switch</a></h3> | ||
| - | <p><span>Cro:<span> Transcription inhibitor. The protein Cro can automatically form diamer Cro2 and then binds OR3, OR2 and OR1 (affinity OR3 > OR2 = OR1, i.e. preferentially binds OR3 | + | <p><span>Cro:<span> Transcription inhibitor. The protein Cro can automatically form diamer Cro2 and then binds OR3, OR2 and OR1 (affinity OR3 > OR2 = OR1, i.e. preferentially binds OR3).At low concentrations the Cro2 blocks the pRM promoter, preventing cI production afterwards. At high concentrations the Cro2 downregulates its own production through OR2 and OR1 binding. No cooperative binding (c.f. below for cI binding)</p> |
<p><span>CI:<span> Transcription inhibitor. The protein CI can also automatically form diamer CI2(similar to Cro2) and then binds OR1, OR2 and OR3 (affinity OR1 > OR2 = OR3, i.e. preferentially binds OR1). At low concentrations the CI2 blocks the pR promoter, preventing cro production afterwards. At high concentrations the CI2 downregulates its own production through OR3 binding. N terminal domain of cI on OR2 tightens the binding of RNA polymerase holoenzyme complex to pRM and hence stimulate its own transcription. Repressor also inhibits transcription from the pL promoter.</p> | <p><span>CI:<span> Transcription inhibitor. The protein CI can also automatically form diamer CI2(similar to Cro2) and then binds OR1, OR2 and OR3 (affinity OR1 > OR2 = OR3, i.e. preferentially binds OR1). At low concentrations the CI2 blocks the pR promoter, preventing cro production afterwards. At high concentrations the CI2 downregulates its own production through OR3 binding. N terminal domain of cI on OR2 tightens the binding of RNA polymerase holoenzyme complex to pRM and hence stimulate its own transcription. Repressor also inhibits transcription from the pL promoter.</p> | ||
<h3><a name="2.3">Maintenance of lysogeny</a></h3> | <h3><a name="2.3">Maintenance of lysogeny</a></h3> | ||
<p>Lysogeny is maintained solely by cI. cI represses transcription from pL and pR while upregulating and controlling its own expression from pRM. It is therefore the only protein expressed by lysogenic phage.</p> | <p>Lysogeny is maintained solely by cI. cI represses transcription from pL and pR while upregulating and controlling its own expression from pRM. It is therefore the only protein expressed by lysogenic phage.</p> | ||
| - | <div class="imgholder1" align=" | + | <div class="imgholder1" align="right" style="float:right;width:800px;height:auto;margin-right:0px;"> |
| - | <a href="https://static.igem.org/mediawiki/2012/ | + | <a href="https://static.igem.org/mediawiki/2012/8/8a/Lysogenic_stage.png"><img src="https://static.igem.org/mediawiki/2012/8/8a/Lysogenic_stage.png" alt="project image" style="clear:both;width:800px;height:267px;"></a><br><p><small align="center">CI dimers bound cooperatively to adjacent operator sites in OR and OL. The CI dimers are shown in blue. Each subunit of the dimer consists of an N-terminal DNA-binding domain (N), a C-terminal oligomerization domain(C), and a linker region (black) connecting the two. The dimer pair bound cooperatively at OR1 and OR2 represses transcription from PR and the dimer bound at OR2 also activates transcription from PRM. The dimer pair bound cooperatively at OL1 and OL2 represses transcription from PL.</small></p> |
</div> | </div> | ||
<h3><a name="2.4">Transformation into Lytic life cycle</a></h3> | <h3><a name="2.4">Transformation into Lytic life cycle</a></h3> | ||
| Line 162: | Line 160: | ||
Cro achieves its function by binding to the same operators as (el) repressor protein. </li> | Cro achieves its function by binding to the same operators as (el) repressor protein. </li> | ||
</ul> | </ul> | ||
| - | <br/> | + | <img src="https://static.igem.org/mediawiki/2012/a/ad/Lytic_stage%28v2.0%29.png" style="clear:both;width:800px;height:267px;"> |
| + | <small align="center">The Cro diamer binds to the OR3, then the transcription from pRM will be blocked. When the concentration of CI drops down, the efficiency of transcription from pR will gradually return normal. | ||
| + | </small> | ||
| + | </br> | ||
| + | </br> | ||
| + | </br> | ||
<h3><a name="2.5">Lytic or lysogenic? </a></h3> | <h3><a name="2.5">Lytic or lysogenic? </a></h3> | ||
<p>The gene regulatory circuitry of phage λ is among the best-understood circuits at the mechanistic level. This circuitry involves several interesting regulatory behaviors. An infected cell undergoes a decision between two alternative pathways, the lytic and lysogenic pathways, just like two states controlled by a toggle switch.</p> | <p>The gene regulatory circuitry of phage λ is among the best-understood circuits at the mechanistic level. This circuitry involves several interesting regulatory behaviors. An infected cell undergoes a decision between two alternative pathways, the lytic and lysogenic pathways, just like two states controlled by a toggle switch.</p> | ||
| Line 179: | Line 182: | ||
| - | <div id="nav_5" class="nav_5style" style="position:fixed; left:5px; top:120px; width:110px; height:430px;background-color: #DFFFDF; -moz-border-radius: 10px; -webkit-border-radius: 10px;border-radius: 10px; border: 1px solid DDD; padding:8px;margin:0px;border:none;"> | + | <div id="nav_5" class="nav_5style" style="position: fixed; left:5px; top:120px; width:110px; height:430px;background-color: #DFFFDF; -moz-border-radius: 10px; -webkit-border-radius: 10px;border-radius: 10px; border: 1px solid DDD; size:auto; padding:8px;margin:0px;border:none;"> |
<p class="level0"><a href="https://2012.igem.org/Team:USTC-China">HOME</a></p> | <p class="level0"><a href="https://2012.igem.org/Team:USTC-China">HOME</a></p> | ||
| Line 189: | Line 192: | ||
<p class="level1"><a href="https://2012.igem.org/Team:USTC-China/methods">Methods</a></p> | <p class="level1"><a href="https://2012.igem.org/Team:USTC-China/methods">Methods</a></p> | ||
| - | <p class="level0"><a href="https://2012.igem.org/Team:USTC-China/ | + | <p class="level0"><a href="https://2012.igem.org/Team:USTC-China/results">ACHIEVEMENTS</a></p> |
<p class="level1"><a href="https://2012.igem.org/Team:USTC-China/results">Results</a></p> | <p class="level1"><a href="https://2012.igem.org/Team:USTC-China/results">Results</a></p> | ||
Latest revision as of 23:31, 26 September 2012
PROJECT BACKGROUND
Phage’s damage towards the fermentation
The phage is a kind of virus that infects bacteria. Nowadays, nearly each kind of prokaryote has been found to have its specific phage. Generally, a phage contains a head, a collar and a tail. The head, namely a protein capsid, contains the phage's genetic material and the tail functions as an anchor for binding to the membrane of the bacteria. The phage is tiny and can only be seen under the EMS (electron microscope). The phage has intense infectivity. What's worse, the phage's resistance against the physical and chemical factors is far too stronger than that of bacteria. It can easily survive the toxic organic solvent such as diethyl ether, the chloroform and the severe circumstance such as low temperature and freeze. However, the phage is sensitive to ultraviolet rays and heat.
The phage widely distributes in the nature, especially in the air around the manufacturing shops of the fermentation factory and the drainage system. The phage can spread by air and sneak into nearly every procedure of the fermentation. Main reasons for the pollution of one single fermentation tank include: remaining blind sides both in the tank and its assistant pipes and the misoperation during inoculating. Main sources of the mass invasion of phages include air filtration system, importation of new strains, lysogen and unthoroughly autoclaved media. All these reasons and sources can lead to continuous pollution among tanks and massive infection.
In fermentation production, the loss that results from the infection by the phage is striking. Once the bacteria are infected, it is very difficult to eradicate the phages and the lysogens thoroughly because of the intense infection and rapid spread. For this reason, it is quite important to prevent the invasion of the phages. If large tanks of fermentation broth are only polluted slightly, the period of the fermentation will be extended, the broth will become clear gradually and the synthesis of the fermented products will be much more difficult. However, if the fermentation broth is unluckily polluted severely, the production has to stop totally, and massive loss in profit happens afterward. This kind of catastrophe occurs commonly in the fermentation of glutamic acid, amylase, protease, acetone and many kinds of antibiotics. For example, in the fermentation of lactic acid, the production of the starter culture of lactobacillus will stop in 30 minutes once the bacteria are infected severely by phages.
Some common phenomena that will occur if the fermentation broth is polluted by the phages are listed below:
Introduction to lambda phage
Entcrobacteria phage λ (lambda phage, coliphage λ) is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. This virus is temperate and may reside within the genome of its host through lysogeny or amplify its number and lyse the host through lytic life cycle.
Usually, a "lytic cycle" ensues, where the lambda DNA is replicated many times and the genes for head, tail and lysis proteins are expressed. This leads to assembly of multiple new phage particles within the cell and subsequent cell lysis, releasing the cell contents, including virions that have been assembled, into the environment
However, under certain conditions the phage DNA may integrate itself into the host cell chromosome in the lysogenic pathway. In this state, the λ DNA is called a prophage and stays resident within the host's genome without apparent harm to the host. The host can be termed a lysogen when a prophage is present.
Genome structure of the toggle switch in lambda phage
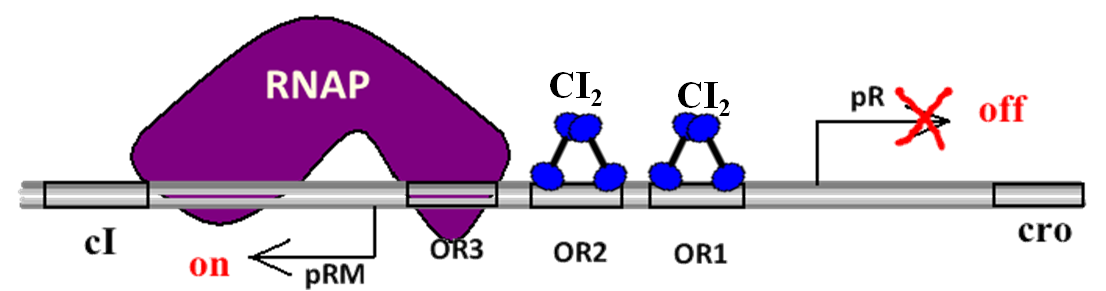
Promoter pR:Initializes the rightward transcription. If the pR is not repressed, these proteins will be expressed and the phage will transfer into lytic cycle and plenty of new phage particles will be assembled.
Promoter pRM:Initializes the transcription of repressor CI. It transcripts leftwards. The transcription efficiency of pRM is quite low, nearly one eighth of the efficiency of pR.
OR1, OR2, OR3: Three protein binding regions on the genome of lambda phage. They can be bound by protein CI dimmers(CI2) and Cro diamers(Cro2). The sites of these three regions on DNA are shown in the picture below.
Function overview of the proteins involve in the toggle switch
Cro: Transcription inhibitor. The protein Cro can automatically form diamer Cro2 and then binds OR3, OR2 and OR1 (affinity OR3 > OR2 = OR1, i.e. preferentially binds OR3).At low concentrations the Cro2 blocks the pRM promoter, preventing cI production afterwards. At high concentrations the Cro2 downregulates its own production through OR2 and OR1 binding. No cooperative binding (c.f. below for cI binding)
CI: Transcription inhibitor. The protein CI can also automatically form diamer CI2(similar to Cro2) and then binds OR1, OR2 and OR3 (affinity OR1 > OR2 = OR3, i.e. preferentially binds OR1). At low concentrations the CI2 blocks the pR promoter, preventing cro production afterwards. At high concentrations the CI2 downregulates its own production through OR3 binding. N terminal domain of cI on OR2 tightens the binding of RNA polymerase holoenzyme complex to pRM and hence stimulate its own transcription. Repressor also inhibits transcription from the pL promoter.
Maintenance of lysogeny
Lysogeny is maintained solely by cI. cI represses transcription from pL and pR while upregulating and controlling its own expression from pRM. It is therefore the only protein expressed by lysogenic phage.
CI dimers bound cooperatively to adjacent operator sites in OR and OL. The CI dimers are shown in blue. Each subunit of the dimer consists of an N-terminal DNA-binding domain (N), a C-terminal oligomerization domain(C), and a linker region (black) connecting the two. The dimer pair bound cooperatively at OR1 and OR2 represses transcription from PR and the dimer bound at OR2 also activates transcription from PRM. The dimer pair bound cooperatively at OL1 and OL2 represses transcription from PL.
Transformation into Lytic life cycle
Cro is responsible for preventing the synthesis of the repressor CI and this action shuts off the possibility of establishing lysogeny. It has two effects:
- It prevents the synthesis of repressor via the maintenance circuit; that is, it prevents transcription via pRM .
- It also inhibits the expression of early genes from both pL and pR Cro achieves its function by binding to the same operators as (el) repressor protein.
 The Cro diamer binds to the OR3, then the transcription from pRM will be blocked. When the concentration of CI drops down, the efficiency of transcription from pR will gradually return normal.
The Cro diamer binds to the OR3, then the transcription from pRM will be blocked. When the concentration of CI drops down, the efficiency of transcription from pR will gradually return normal.
Lytic or lysogenic?
The gene regulatory circuitry of phage λ is among the best-understood circuits at the mechanistic level. This circuitry involves several interesting regulatory behaviors. An infected cell undergoes a decision between two alternative pathways, the lytic and lysogenic pathways, just like two states controlled by a toggle switch.
Simplistically, in cells with sufficient nutrients, protease activity is high, which breaks down cI.[10] This leads to the lytic lifestyle. In cells with limited nutrients, protease activity is low, making cI stable. This leads to the lysogenic lifestyle. This means that a cell "in trouble", i.e. lacking in nutrients and in a more dormant state, is more likely to lysogenise. This would be selected for because the phage can now lie dormant in the bacterium until it falls on better times, and so the phage can create more copies of itself with the additional resources available and with the more likely proximity of further infectable cells.
 "
"





